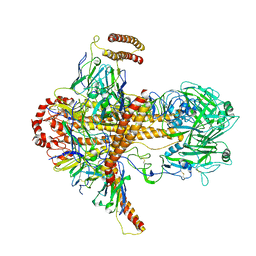
7DKJ
 
 | |
2KHE
 
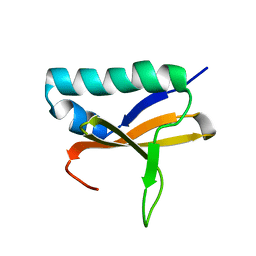 | | Solution Structure of the Bacterial Toxin Rele from Thermus Thermophilus HB8 | | Descriptor: | Toxin-like protein | | Authors: | Suzuki, S, Kawazoe, M, Kaminishi, T, Takemoto, C, Muto, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacterial Toxin Rele from Thermus Thermophilus HB8
To be Published
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | Descriptor: | SOS 1 | | Authors: | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-02-18 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
4YN3
 
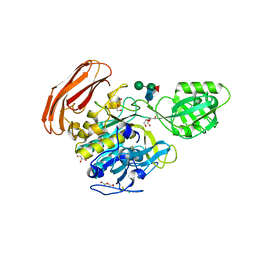 | | Crystal structure of Cucumisin complex with pro-peptide | | Descriptor: | CHLORIDE ION, Cucumisin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murayama, K, Kato-Murayama, M, Yokoyama, S, Arima, K, Shirouzu, M. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of cucumisin protease activity regulation by its propeptide
J. Biochem., 161, 2017
|
|
8XM7
 
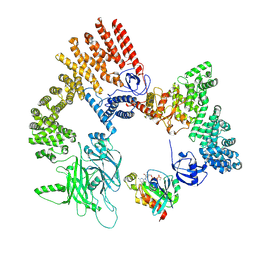 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex: RhoG/DOCK5/ELMO1 focused map | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-12-27 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.91 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
9J0P
 
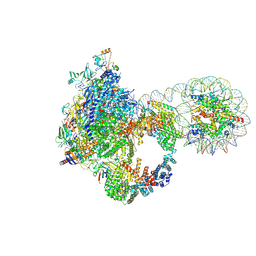 | | Arrested elongation complex of mammalian RNA polymerase II with nucleosome (AEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
9J0N
 
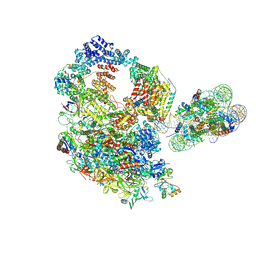 | | Paused elongation complex of mammalian RNA polymerase II with nucleosome (PEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
9J0O
 
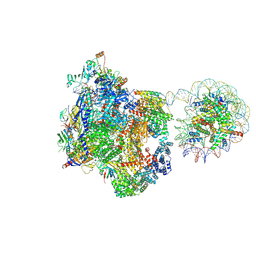 | | Arrested elongation complex of mammalian RNA polymerase II with nucleosome (AEC1-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
8B64
 
 | | Cryo-EM structure of RC-LH1-PufX photosynthetic core complex from Rba. capsulatus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.589 Å) | | Cite: | Cryo-EM structure of a monomeric RC-LH1-PufX supercomplex with high-carotenoid content from Rhodobacter capsulatus.
Structure, 31, 2023
|
|
5XWD
 
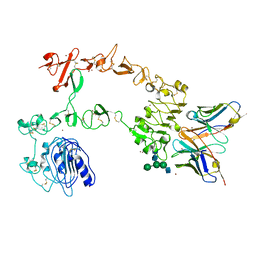 | | Crystal structure of the complex of 059-152-Fv and EGFR-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Matsuda, T, Ito, T, Shirouzu, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction
PLoS ONE, 13, 2018
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
8W9G
 
 | | Hepatitis B virus core protein Y132A mutant in complex with CBT-078 | | Descriptor: | 2-fluoranyl-N1-(4-fluorophenyl)-N3-(2-methylphenyl)benzene-1,3-dicarboxamide, Capsid protein | | Authors: | Iwasaki, W, Katsura, K, Tomabechi, Y, Niwa, H, Ogawa, K, Kojima, S, Shirouzu, M. | | Deposit date: | 2023-09-05 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Screening systems for the detection of hepatitis B virus replication and capsid assembly
To Be Published
|
|
9KLB
 
 | | G9a in complex with RK-133232 (compound 16g) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of potent substrate-type lysine methyltransferase G9a inhibitors for the treatment of sickle cell disease.
Eur.J.Med.Chem., 293, 2025
|
|
9KLC
 
 | | G9a in complex with the S-isomer of RK-131902 (racemic compound rac-10a) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of potent substrate-type lysine methyltransferase G9a inhibitors for the treatment of sickle cell disease.
Eur.J.Med.Chem., 293, 2025
|
|
5XOG
 
 | | RNA Polymerase II elongation complex bound with Spt5 KOW5 and Elf1 | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (30-MER), DNA (39-MER), ... | | Authors: | Ehara, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
5H09
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-ethyl2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoate | | Descriptor: | Tyrosine-protein kinase HCK, ethyl (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanoate | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0B
 
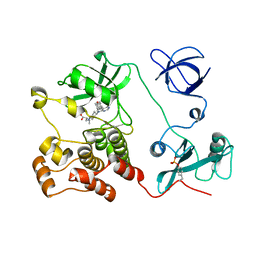 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
1ZRJ
 
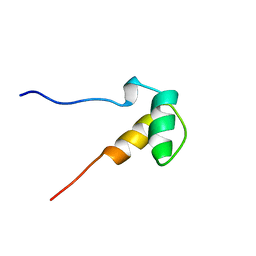 | | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c | | Descriptor: | E1B-55kDa-associated protein 5 isoform c | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c
To be Published
|
|
1Z54
 
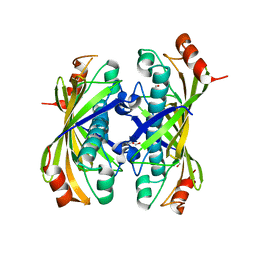 | | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus | | Descriptor: | GLYCEROL, probable thioesterase | | Authors: | Ihsanawati, Kaminishi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus
TO BE PUBLISHED
|
|
1ZPW
 
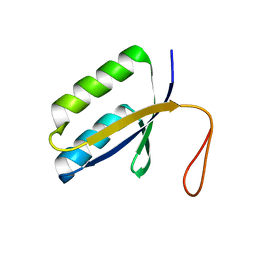 | |
3B13
 
 | | Crystal structure of the DHR-2 domain of DOCK2 in complex with Rac1 (T17N mutant) | | Descriptor: | Dedicator of cytokinesis protein 2, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Mishima-Tsumagari, C, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-06-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WR5
 
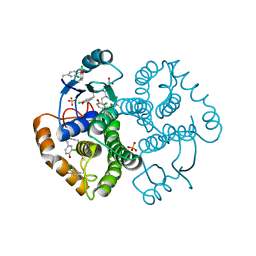 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WR4
 
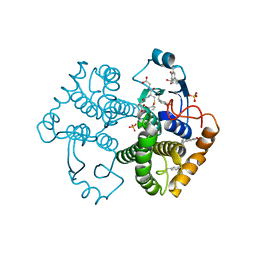 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
8ZJJ
 
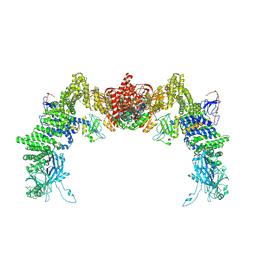 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 2) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJL
 
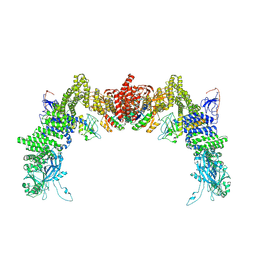 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 4) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
