1WR0
 
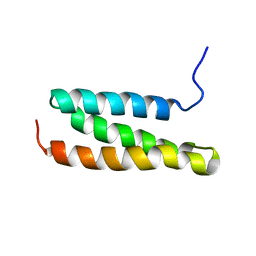 | | Structural characterization of the MIT domain from human Vps4b | | Descriptor: | SKD1 protein | | Authors: | Takasu, H, Jee, J.G, Ohno, A, Goda, N, Fujiwara, K, Tochio, H, Shirakawa, M, Hiroaki, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-07 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the MIT domain from human Vps4b
Biochem.Biophys.Res.Commun., 334, 2005
|
|
1GEA
 
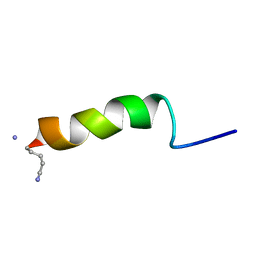 | | RECEPTOR-BOUND CONFORMATION OF PACAP21 | | Descriptor: | PITUITARY ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE | | Authors: | Inooka, H, Ohtaki, T, Kitahara, O, Ikegami, T, Endo, S, Kitada, C, Ogi, K, Onda, H, Fujino, M, Shirakawa, M. | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Conformation of a peptide ligand bound to its G-protein coupled receptor.
Nat.Struct.Biol., 8, 2001
|
|
1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
1D9N
 
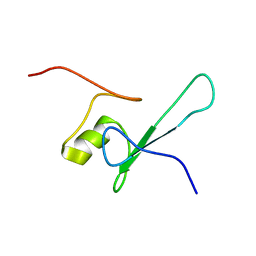 | | SOLUTION STRUCTURE OF THE METHYL-CPG-BINDING DOMAIN OF THE METHYLATION-DEPENDENT TRANSCRIPTIONAL REPRESSOR MBD1/PCM1 | | Descriptor: | METHYL-CPG-BINDING PROTEIN MBD1 | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Nakao, M, Shirakawa, M. | | Deposit date: | 1999-10-28 | | Release date: | 2000-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1.
EMBO J., 18, 1999
|
|
1WLF
 
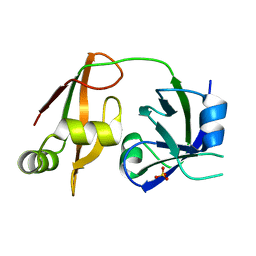 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
2RRE
 
 | | Structure and function of the N-terminal nucleolin binding domain of nuclear valocine containing protein like 2 (NVL2) harboring a nucleolar localization signal | | Descriptor: | Putative uncharacterized protein | | Authors: | Fujiwara, Y, Fujiwara, K, Goda, N, Iwaya, N, Tenno, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2010-08-03 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal nucleolin binding domain of nuclear valosin-containing protein-like 2 (NVL2) harboring a nucleolar localization signal
J.Biol.Chem., 286, 2011
|
|
1FR0
 
 | | SOLUTION STRUCTURE OF THE HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ANAEROBIC SENSOR KINASE ARCB FROM ESCHERICHIA COLI. | | Descriptor: | ARCB | | Authors: | Ikegami, T, Okada, T, Ohki, I, Hirayama, J, Mizuno, T, Shirakawa, M. | | Deposit date: | 2000-09-07 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamic character of the histidine-containing phosphotransfer domain of anaerobic sensor kinase ArcB from Escherichia coli.
Biochemistry, 40, 2001
|
|
1COP
 
 | |
1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
2ROG
 
 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
1IRY
 
 | | Solution structure of the hMTH1, a nucleotide pool sanitization enzyme | | Descriptor: | hMTH1 | | Authors: | Mishima, M, Itoh, N, Sakai, Y, Kamiya, H, Nakabeppu, Y, Shirakawa, M. | | Deposit date: | 2001-10-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates
J.Biol.Chem., 279, 2004
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
1KLV
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor associated protein | | Descriptor: | GABA(A) Receptor associated protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-13 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
1KM7
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor Associated Protein | | Descriptor: | GABA(A) Receptor Associated Protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-14 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
3VYB
 
 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/hmCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5HC)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VXX
 
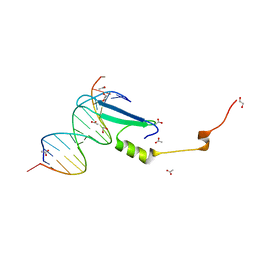 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/5mCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5CM)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYQ
 
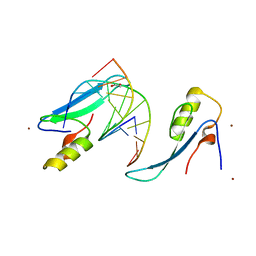 | | Crystal structure of the methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence in space group P1 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*AP*TP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VXV
 
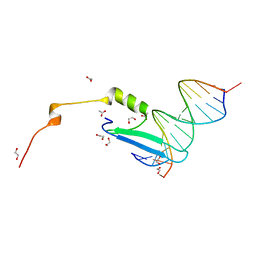 | | Crystal structure of methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*CP*(5CM)P*GP*GP*AP*CP*A)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
3WO4
 
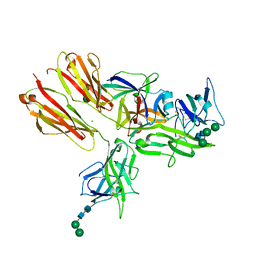 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO2
 
 | | Crystal structure of human interleukin-18 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO3
 
 | | Crystal structure of IL-18 in complex with IL-18 receptor alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
1J0S
 
 | | Solution structure of the human interleukin-18 | | Descriptor: | Interleukin-18 | | Authors: | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
2RR9
 
 | | The solution structure of the K63-Ub2:tUIMs complex | | Descriptor: | Putative uncharacterized protein UIMC1, ubiquitin | | Authors: | Sekiyama, N, Jee, J, Isogai, S, Akagi, K, Huang, T, Ariyoshi, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the K63-Ub2:tUIMs complex
To be Published
|
|
2RPQ
 
 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
