1A0A
 
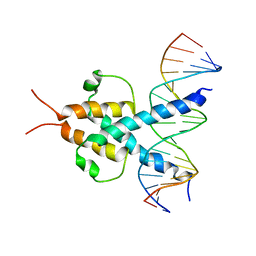 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
2MLQ
 
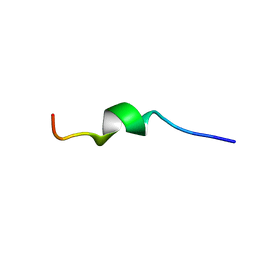 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
3WZU
 
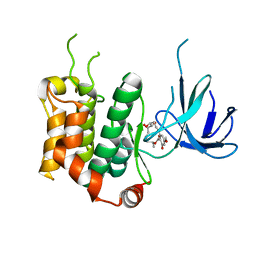 | | THE STRUCTURE OF MAP2K7 IN COMPLEX WITH 5Z-7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, Y, Matsumoto, T, Kinoshita, T. | | Deposit date: | 2014-10-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | 5Z-7-Oxozeaenol covalently binds to MAP2K7 at Cys218 in an unprecedented manner.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3WZQ
 
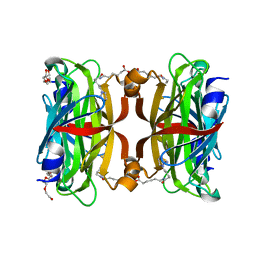 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZP
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X00
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
7Q5V
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
5ZX8
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Matsumoto, A, Uehara, U, Shimizu, Y, Ueda, T, Uchiumi, T, Ito, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus.
Proteins, 87, 2019
|
|
3WZN
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin at 1.3 A resolution | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
3WZO
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin long tail (BTNtail) at 1.5 A resolution | | Descriptor: | 6-({5-[(3aS,4S,5S,6aR)-5-oxido-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, CADMIUM ION, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
7Q5X
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH 2-OXOGLUTARATE (2OG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
8YFY
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 138, 2024
|
|
8YFZ
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
1BA5
 
 | | DNA-BINDING DOMAIN OF HUMAN TELOMERIC PROTEIN, HTRF1, NMR, 18 STRUCTURES | | Descriptor: | HTRF1 | | Authors: | Nishikawa, T, Nagadoi, A, Yoshimura, S, Aimoto, S, Nishimura, Y. | | Deposit date: | 1998-04-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of human telomeric protein, hTRF1.
Structure, 6, 1998
|
|
1UBB
 
 | | Crystal structure of rat HO-1 in complex with ferrous heme | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Higashimoto, Y, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
3WKT
 
 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J3Z
 
 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
2RSO
 
 | |
2RSN
 
 | |
8Z8R
 
 | |
8ZFW
 
 | |
8Z8S
 
 | |
8Z8P
 
 | |
8Z8O
 
 | |
