7EAY
 
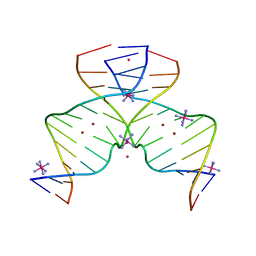 | | DNA containing Cu(II)-mediated 4-N-carboxymethylcytosine base pairs | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), COPPER (II) ION, ... | | Authors: | Kondo, J, Terashima, A, Yoshimura, A, Tada, Y, Ono, A. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA containing Cu(II)-mediated 4-N-carboxymethylcytosine base pairs
To Be Published
|
|
5XDX
 
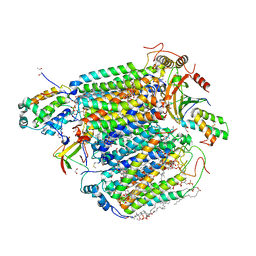 | | Bovine heart cytochrome c oxidase in the reduced state with pH 7.3 at 1.99 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-03-30 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of bovine cytochrome c oxidase in the ligand-free reduced state at neutral pH.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5XDQ
 
 | | Bovine heart cytochrome c oxidase in the fully oxidized state with pH 7.3 at 1.77 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
2EH9
 
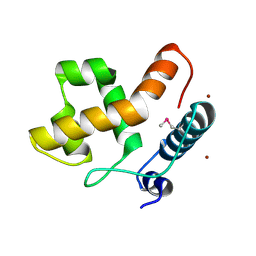 | |
8GVM
 
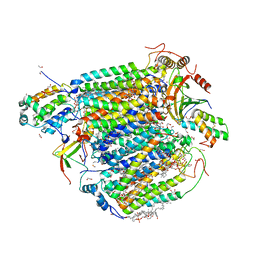 | | The structure of azide-bound cytochrome C oxidase determined using the crystals exposed to 20 mm azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity
J. Biol. Chem., 293, 2018
|
|
6KFG
 
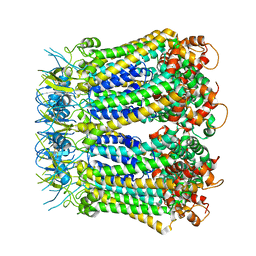 | | Undocked INX-6 hemichannel in detergent | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
2E1M
 
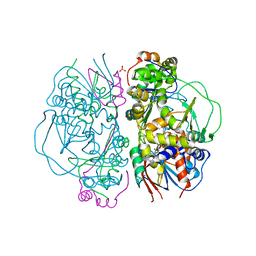 | | Crystal Structure of L-Glutamate Oxidase from Streptomyces sp. X-119-6 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, PHOSPHATE ION | | Authors: | Sasaki, C, Kashima, A, Sakaguchi, C, Mizuno, H, Arima, J, Kusakabe, H, Tamura, T, Sugio, S, Inagaki, K. | | Deposit date: | 2006-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6
Febs J., 276, 2009
|
|
8IJN
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
1BFG
 
 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
6IKO
 
 | | Crystal structure of mouse GAS7cb | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.756 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
3WAK
 
 | |
2PT5
 
 | | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Shikimate kinase | | Authors: | Jeyakanthan, J, Nithya, N, Shimada, A, Velmurugan, D, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5
To be Published
|
|
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
7WSV
 
 | | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | | Descriptor: | Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2ZDU
 
 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(4-{[6-bromo-3-(methoxycarbonyl)-1-oxo-4-phenylisoquinolin-2(1H)-yl]methyl}phenyl)amino]-4-oxobutanoic acid, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Ohra, T, Itoh, F, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (1)
Bioorg.Med.Chem., 16, 2008
|
|
6K7D
 
 | |
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | Descriptor: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
1IYW
 
 | | Preliminary Structure of Thermus thermophilus Ligand-Free Valyl-tRNA Synthetase | | Descriptor: | Valyl-tRNA Synthetase | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-09-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
3WOB
 
 | | Crystal structure of a prostate-specific WGA16 glycoprotein lectin, form I | | Descriptor: | hypothetical protein | | Authors: | Garenaux, E, Kanagawa, M, Tsuchiyama, T, Hori, K, Kanazawa, T, Goshima, A, Chiba, M, Yasue, H, Ikeda, A, Yamaguchi, Y, Sato, C, Kitajima, K. | | Deposit date: | 2013-12-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery, Primary, and Crystal Structures and Capacitation-related Properties of a Prostate-derived Heparin-binding Protein WGA16 from Boar Sperm
J.Biol.Chem., 290, 2015
|
|
3WAJ
 
 | |
6K7F
 
 | | Crystal structure of MBPholo-Tim21 fusion protein with a 17-residue helical linker | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Mitochondrial import inner membrane translocase subunit TIM21, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bala, S, Shimada, A, Kohda, D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6IKN
 
 | | Crystal structure of the GAS7 F-BAR domain | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
