2E8A
 
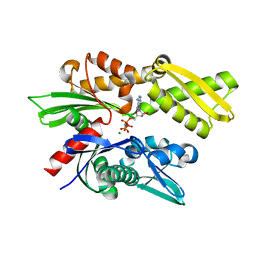 | | Crystal structure of the human Hsp70 ATPase domain in complex with AMP-PNP | | Descriptor: | Heat shock 70kDa protein 1A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2E88
 
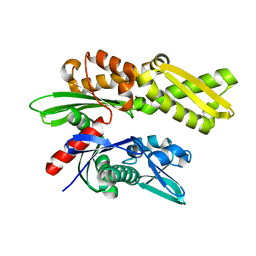 | | Crystal structure of the human Hsp70 ATPase domain in the apo form | | Descriptor: | Heat shock 70kDa protein 1A, ZINC ION | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1ULK
 
 | | Crystal Structure of Pokeweed Lectin-C | | Descriptor: | lectin-C | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
1ULM
 
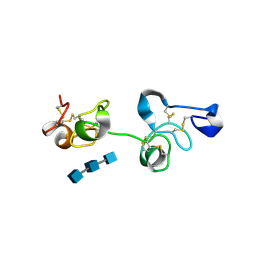 | | Crystal Structure of Pokeweed Lectin-D2 complexed with tri-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin-D2 | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
8YTN
 
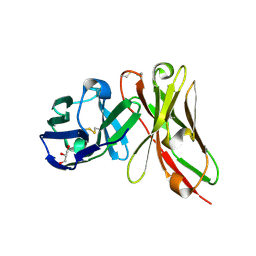 | | Single-chain Fv antibody of E11 | | Descriptor: | GLYCEROL, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTO
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTP
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine under reducing conditions | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
1A0F
 
 | | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTATHIONESULFONIC ACID | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID | | Authors: | Nishida, M, Harada, S, Noguchi, S, Inoue, H, Takahashi, K, Satow, Y. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106.
J.Mol.Biol., 281, 1998
|
|
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
7W5O
 
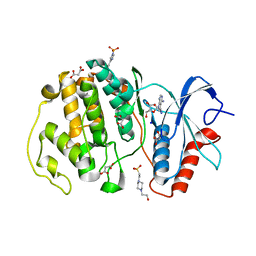 | | Crystal structure of ERK2 with an allosteric inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a novel target site for ATP-independent ERK2 inhibitors.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
2QKS
 
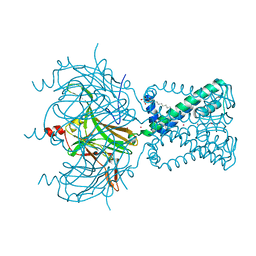 | |
7XC1
 
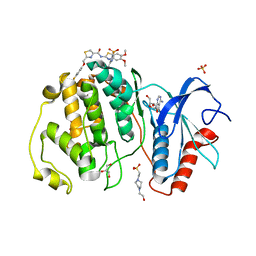 | | Crystal structure of ERK2 with an allosteric inhibitor 3 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for ERK2 allosteric inhibitors.
To Be Published
|
|
7X4U
 
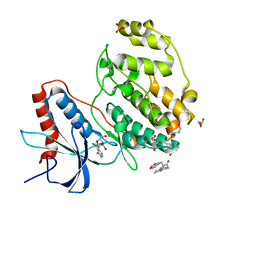 | | Crystal structure of ERK2 with an allosteric inhibitor 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for producing allosteric ERK2 inhibitors
To Be Published
|
|
1N9P
 
 | |
1GCP
 
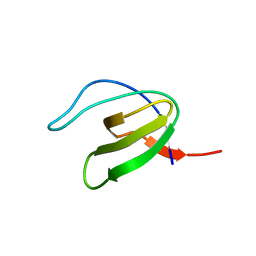 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
2ZC8
 
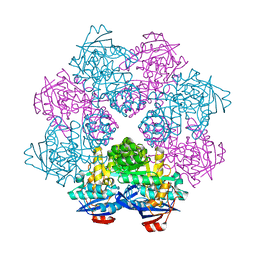 | | Crystal structure of N-Acylamino Acid Racemase from Thermus thermophilus HB8 | | Descriptor: | N-acylamino acid racemase | | Authors: | Hayashida, M, Kim, S.H, Takeda, K, Hisano, T, Miki, K. | | Deposit date: | 2007-11-05 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-acylamino acid racemase from Thermus thermophilus HB8
Proteins, 71, 2008
|
|
4V4O
 
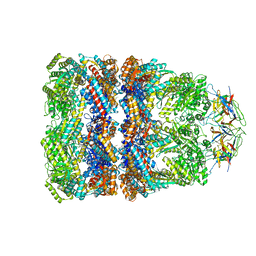 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
5Y0Z
 
 | | Human SIRT2 in complex with a specific inhibitor, NPD11033 | | Descriptor: | (1~{R},9~{S})-11-[(2~{R})-3-[2,4-bis(2-methylbutan-2-yl)phenoxy]-2-oxidanyl-propyl]-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-07-19 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a novel small molecule that inhibits deacetylase but not defatty-acylase reaction catalysed by SIRT2.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
3APD
 
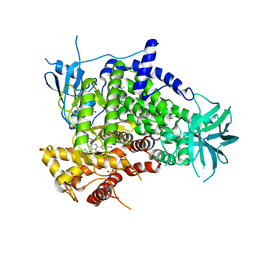 | | Crystal structure of human PI3K-gamma in complex with CH5108134 | | Descriptor: | 5-(2-Morpholin-4-yl-7-pyridin-3-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3APF
 
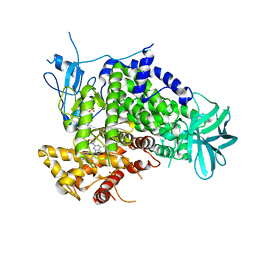 | | Crystal structure of human PI3K-gamma in complex with CH5039699 | | Descriptor: | 3-[7-(1H-benzimidazol-5-yl)-2-(morpholin-4-yl)-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl]phenol, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7PCE
 
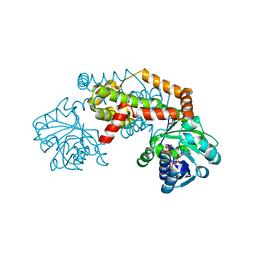 | | BurG (apo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketol-acid reductoisomerase, PHOSPHATE ION | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCO
 
 | | BurG E232Q mutant (holo) in complex with 2R,3R-2,3-dihydroxy-6-methyl-heptanoate (12): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R,3R)-6-methyl-2,3-bis(oxidanyl)heptanoic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCT
 
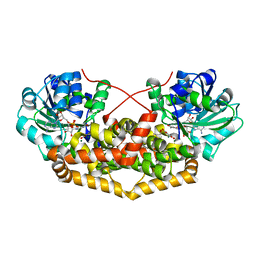 | | BurG E232Q mutant (holo) in complex with enol-oxalacetate (15): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCM
 
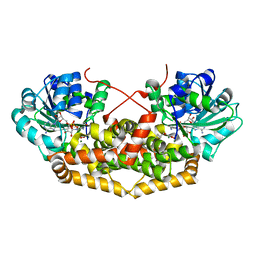 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCC
 
 | | BurG in complex with Mg2+ and NAD+ (holo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
