4EBW
 
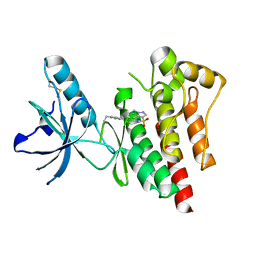 | | Structure of Focal Adhesion Kinase catalytic domain in complex with novel allosteric inhibitor | | Descriptor: | 1-ethyl-8-(4-ethylphenyl)-5-methyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazine 4,4-dioxide, Focal adhesion kinase 1 | | Authors: | Iwatani, M, Iwata, H, Okabe, A, Skene, R.J, Tomita, N, Hayashi, Y, Aramaki, Y, Hosfield, D.J, Hori, A, Baba, A, Miki, H. | | Deposit date: | 2012-03-25 | | Release date: | 2012-07-25 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel allosteric FAK inhibitors.
Eur.J.Med.Chem., 61, 2013
|
|
3B4R
 
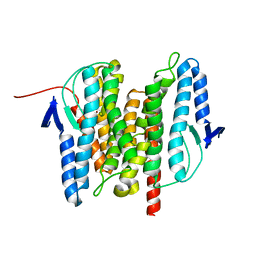 | | Site-2 Protease from Methanocaldococcus jannaschii | | Descriptor: | Putative zinc metalloprotease MJ0392, ZINC ION | | Authors: | Jeffrey, P.D, Feng, L, Yan, H, Wu, Z, Yan, N, Wang, Z, Shi, Y. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a site-2 protease family intramembrane metalloprotease.
Science, 318, 2007
|
|
1AN9
 
 | | D-AMINO ACID OXIDASE COMPLEX WITH O-AMINOBENZOATE | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Miura, R, Setoyama, C, Nishina, Y, Shiga, K, Mizutani, H, Miyahara, I, Hirotsu, K. | | Deposit date: | 1997-06-28 | | Release date: | 1997-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic studies on D-amino acid oxidase x substrate complex: implications of the crystal structure of enzyme x substrate analog complex.
J.Biochem.(Tokyo), 122, 1997
|
|
5AUQ
 
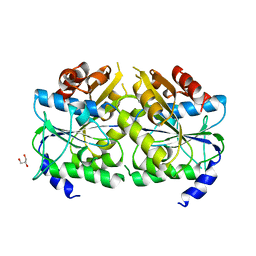 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5B40
 
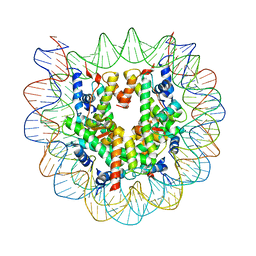 | | The nucleosome structure containing H2B-K120 and H4-K31 monoubiquitinations | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Machida, S, Sekine, S, Nishiyama, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Monoubiquitination of histones H2B and H4 changes the nucleosome stability without affecting the nucleosome structure
To Be Published
|
|
1EVI
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE PURPLE INTERMEDIATE OF PORCINE KIDNEY D-AMINO ACID OXIDASE | | Descriptor: | 3,4-DIHYDRO-2H-PYRROLIUM-5-CARBOXYLATE, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mizutani, H, Miyahara, I, Hirotsu, K, Nishina, Y, Shiga, K, Setoyama, C, Miura, R. | | Deposit date: | 2000-04-20 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of the purple intermediate of porcine kidney D-amino acid oxidase. Optimization of the oxidative half-reaction through alignment of the product with reduced flavin.
J.Biochem.(Tokyo), 128, 2000
|
|
5AUO
 
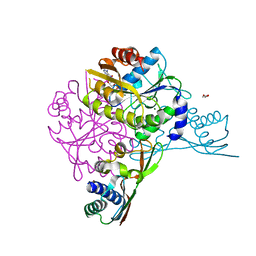 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5B71
 
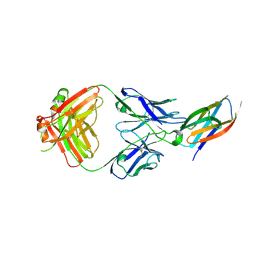 | | Crystal structure of complement C5 in complex with SKY59 | | Descriptor: | Complement C5 beta chain, SKY59 Fab heavy chain, SKY59 Fab light chain | | Authors: | Irie, M, Shimizu, Y, Sampei, Z, Fukuzawa, T. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases.
Sci Rep, 7, 2017
|
|
4XFP
 
 | | Crystal Structure of Highly Active Mutant of Bacillus sp. TB-90 Urate Oxidase | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Hibi, T, Hayashi, Y, Kawamura, A, Itoh, T. | | Deposit date: | 2014-12-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Glycine Substitution of Surface Proline 287 Involves Entropic Enhancement of Bacillus sp. TB-90 Uricase Activity
To be published
|
|
5BCA
 
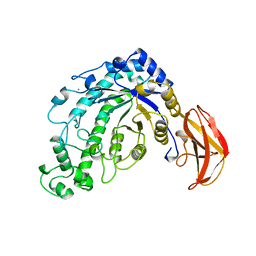 | | BETA-AMYLASE FROM BACILLUS CEREUS VAR. MYCOIDES | | Descriptor: | CALCIUM ION, PROTEIN (1,4-ALPHA-D-GLUCAN MALTOHYDROLASE.) | | Authors: | Oyama, T, Kusunoki, M, Kishimoto, Y, Takasaki, Y, Nitta, Y. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-amylase from Bacillus cereus var. mycoides at 2.2 A resolution.
J.Biochem.(Tokyo), 125, 1999
|
|
5C5O
 
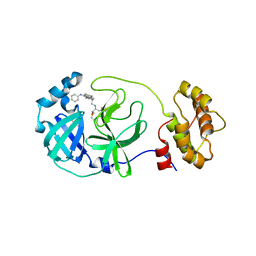 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
8J3S
 
 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
1UFZ
 
 | | Solution structure of HBS1-like domain in hypothetical protein BAB28515 | | Descriptor: | Hypothetical protein BAB28515 | | Authors: | He, F, Muto, Y, Hayami, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HBS1-like domain in hypothetical protein BAB28515
To be Published
|
|
7Q5X
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH 2-OXOGLUTARATE (2OG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
1UG8
 
 | | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease | | Descriptor: | Poly(A)-specific Ribonuclease | | Authors: | Nagata, T, Muto, Y, Hayami, N, Uda, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease
To be Published
|
|
7Q5V
 
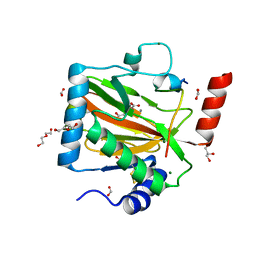 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
1UH6
 
 | | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06 | | Descriptor: | ubiquitin-like 5 | | Authors: | Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-25 | | Release date: | 2003-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06
To be Published
|
|
5CPG
 
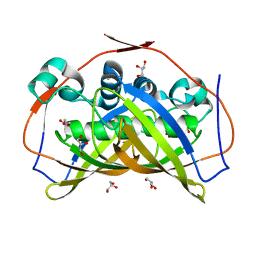 | | R-Hydratase PhaJ1 from Pseudomonas aeruginosa in the unliganded form | | Descriptor: | (R)-specific enoyl-CoA hydratase, GLYCEROL | | Authors: | Tsuge, T, Sato, S, Hiroe, A, Ishizuka, K, Kanazawa, H, Kanagarajan, S, Shiro, Y, Hisano, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Contribution of the Distal Pocket Residue to the Acyl-Chain-Length Specificity of (R)-Specific Enoyl-Coenzyme A Hydratases from Pseudomonas spp.
Appl.Environ.Microbiol., 81, 2015
|
|
1MBF
 
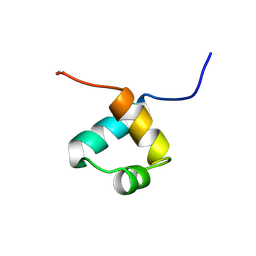 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBH
 
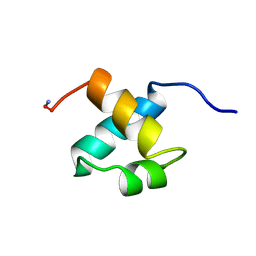 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1O5P
 
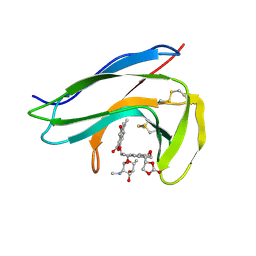 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5AUP
 
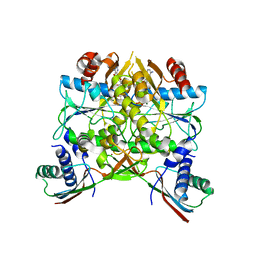 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
