4TWW
 
 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-07-02 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
5X93
 
 | | Human endothelin receptor type-B in complex with antagonist K-8794 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[6-[(4-tert-butylphenyl)sulfonylamino]-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]oxy-N-(2,6-dimethylphenyl)propanamide, CHOLESTEROL, ... | | 著者 | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | 登録日 | 2017-03-05 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
5XPR
 
 | | Human endothelin receptor type-B in complex with antagonist bosentan | | 分子名称: | 4-tert-butyl-N-[6-(2-hydroxyethyloxy)-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]benzenesulfonamide, Endothelin B receptor,Endolysin,Endothelin B receptor, SULFATE ION | | 著者 | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | 登録日 | 2017-06-04 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
8I53
 
 | |
6IMY
 
 | | Crystal structure of V30M mutated transthyretin in complex with 4'-caroboxybenzo-18-Crown-6 | | 分子名称: | 2,3,5,6,8,9,11,12,14,15-decahydro-1,4,7,10,13,16-benzohexaoxacyclooctadecine-18-carboxylic acid, Transthyretin | | 著者 | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | 登録日 | 2018-10-24 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Crown Ethers as Transthyretin Amyloidogenesis Inhibitors.
J. Med. Chem., 62, 2019
|
|
1UIL
 
 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | 分子名称: | Double-stranded RNA-binding motif | | 著者 | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-17 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
1UG0
 
 | | Solution structure of SURP domain in BAB30904 | | 分子名称: | splicing factor 4 | | 著者 | He, F, Muto, Y, Ushikoshi, R, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-11 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of SURP domain in BAB30904
To be Published
|
|
4U4P
 
 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | 分子名称: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | 著者 | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | 登録日 | 2014-07-24 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
8JT7
 
 | | Structure of arginine oxidase from Pseudomonas sp. TRU 7192 | | 分子名称: | Amine oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Yamaguchi, H, Numoto, N, Suzuki, H, Nishikawa, K, Kamegawa, A, Takahashi, K, Sugiki, M, Fujiyoshi, Y. | | 登録日 | 2023-06-21 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.34 Å) | | 主引用文献 | Structural basis of arginine oxidase from Pseudomonas sp. TRU 7192
To Be Published
|
|
8JS7
 
 | | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme | | 分子名称: | GLYCEROL, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | 登録日 | 2023-06-19 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8JS5
 
 | | Dimeric PAS domains of oxygen sensor FixL with ferric unliganded heme | | 分子名称: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | 著者 | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | 登録日 | 2023-06-19 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8JS6
 
 | | Dimeric PAS domains of oxygen sensor FixL in complex with cyanide-bound ferric heme | | 分子名称: | CYANIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | 登録日 | 2023-06-19 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
6IOP
 
 | | The ligand binding domain of Mlp24 | | 分子名称: | ACETATE ION, ALANINE, CALCIUM ION, ... | | 著者 | Sumita, K, Takahashi, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2018-10-31 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
4WBG
 
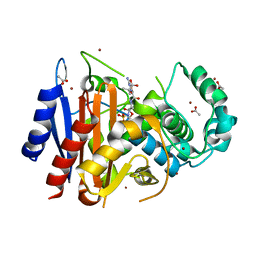 | | Crystal structure of class C beta-lactamase Mox-1 covalently complexed with aztorenam | | 分子名称: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, ACETATE ION, Beta-lactamase, ... | | 著者 | Oguri, T, Shimizu-ibuka, A, Ishii, Y. | | 登録日 | 2014-09-03 | | 公開日 | 2015-07-01 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational Change Observed in the Active Site of Class C beta-Lactamase MOX-1 upon Binding to Aztreonam
Antimicrob.Agents Chemother., 59, 2015
|
|
1UFN
 
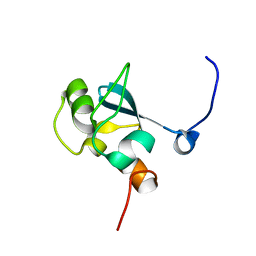 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | 分子名称: | putative nuclear protein homolog 5830484A20Rik | | 著者 | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-02 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | 著者 | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5AZ2
 
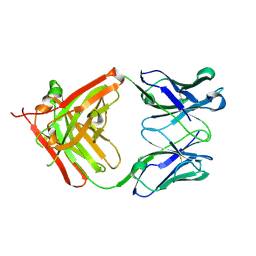 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | 分子名称: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | 著者 | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | 登録日 | 2015-09-16 | | 公開日 | 2015-12-09 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.603 Å) | | 主引用文献 | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
3FHP
 
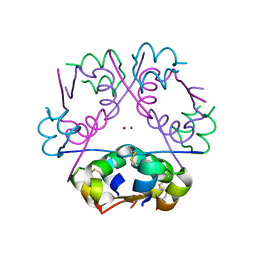 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | 分子名称: | Insulin, ZINC ION | | 著者 | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | 登録日 | 2008-12-09 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å) | | 主引用文献 | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | 分子名称: | tau-AnmTx Ueq 12-1 | | 著者 | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | 登録日 | 2016-06-14 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
4WY3
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-11-15 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4BGN
 
 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | 分子名称: | VOLTAGE-GATED SODIUM CHANNEL | | 著者 | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | 登録日 | 2013-03-28 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (9 Å) | | 主引用文献 | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
8JLB
 
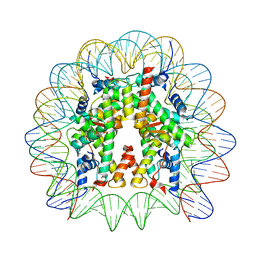 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-02 | | 公開日 | 2023-10-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | 分子名称: | Beta-glucosidase, GLYCEROL | | 著者 | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | 登録日 | 2023-05-03 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
