1DDZ
 
 | | X-RAY STRUCTURE OF A BETA-CARBONIC ANHYDRASE FROM THE RED ALGA, PORPHYRIDIUM PURPUREUM R-1 | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Mitsuhashi, S, Mizushima, T, Yamashita, E, Miyachi, S, Tsukihara, T. | | Deposit date: | 1999-11-12 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO(2) hydration.
J.Biol.Chem., 275, 2000
|
|
4JWK
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Acinetobacter baumannii with cytidine at 1.87 A resolution | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
1J30
 
 | | The crystal structure of sulerythrin, a rubrerythrin-like protein from a strictly aerobic and thermoacidiphilic archaeon | | Descriptor: | 144aa long hypothetical rubrerythrin, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-01-16 | | Release date: | 2003-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Sulerythrin, A Rubrerythrin-like Protein from A Strictly Aerobic Archaeon, Sulfolobus tokodaii strain 7, shows unexpected domain swapping
Biochemistry, 42, 2003
|
|
2RQP
 
 | |
4FOP
 
 | | Crystal Structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.86 A resolution | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kaushik, S, Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
1H6L
 
 | |
1GS3
 
 | |
3QRC
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis, in complex with the heparin analogue sucrose octasulfate | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
3NJU
 
 | | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Kaushik, S, Prem Kumar, R, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution
To be Published
|
|
3QRA
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
4JX9
 
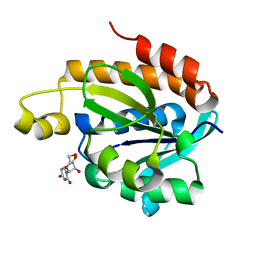 | | Crystal structure of the complex of peptidyl t-RNA hydrolase from Acinetobacter baumannii with uridine at 1.4A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, URIDINE | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-28 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
3RK8
 
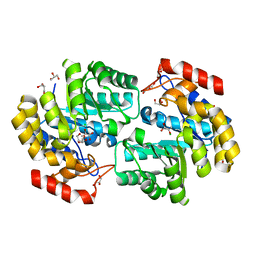 | | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kaushik, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-17 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution
To be Published
|
|
1WDE
 
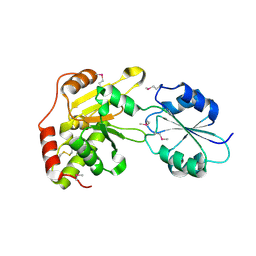 | | Crystal structure of the conserved hypothetical protein APE0931 from Aeropyrum pernix K1 | | Descriptor: | Probable diphthine synthase | | Authors: | Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two archaeal diphthine synthases: insights into the post-translational modification of elongation factor 2.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1WG8
 
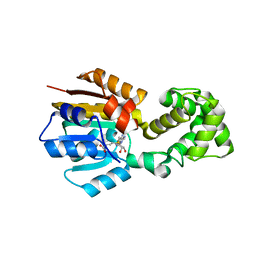 | |
8H4V
 
 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
8HB5
 
 | | Crystal structure of Mincle in complex with HD-275 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-27 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
4LWQ
 
 | | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution
To be Published
|
|
1WX6
 
 | | Solution structure of the SH3 domain of the human cytoplasmic protein NCK2 | | Descriptor: | Cytoplasmic protein NCK2 | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the human cytoplasmic protein NCK2
To be Published
|
|
2IRZ
 
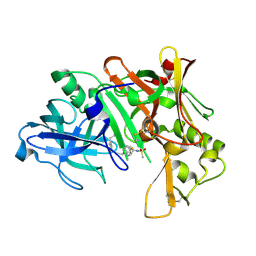 | | Crystal structure of human Beta-secretase complexed with inhibitor | | Descriptor: | 3-{5-[(1R)-1-AMINO-1-METHYL-2-PHENYLETHYL]-1,3,4-OXADIAZOL-2-YL}-N-[(1R)-1-(4-FLUOROPHENYL)ETHYL]-5-[METHYL(METHYLSULFONYL)AMINO]BENZAMIDE, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of oxadiazoyl tertiary carbinamine inhibitors of beta-secretase (BACE-1).
J.Med.Chem., 49, 2006
|
|
2IS0
 
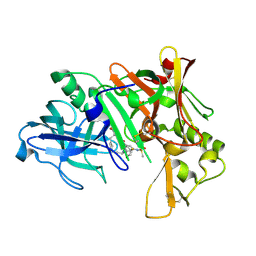 | | Crystal structure of human Beta-secretase complexed with inhibitor | | Descriptor: | (2S)-2-AMINO-2-BENZYL-3-HYDROXYPROPYL 3-({[(1R)-1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)-5-[METHYL(METHYLSULFONYL)AMINO]BENZOATE, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of oxadiazoyl tertiary carbinamine inhibitors of beta-secretase (BACE-1).
J.Med.Chem., 49, 2006
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | Descriptor: | SOS 1 | | Authors: | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-02-18 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
6IF6
 
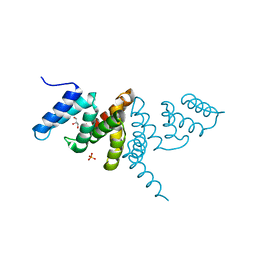 | | Structure of the periplasmic domain of SflA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein SflA | | Authors: | Nishikawa, S, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the periplasmic domain of SflA involved in spatial regulation of the flagellar biogenesis of Vibrio reveals a TPR/SLR-like fold.
J.Biochem., 166, 2019
|
|
2QZK
 
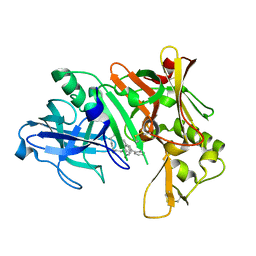 | | Crystal structure of human Beta Secretase complexed with I21 | | Descriptor: | 2-[(5R)-5-amino-5-methyl-4,16-dioxo-14-phenyl-3-oxa-15-azatricyclo[15.3.1.1~7,11~]docosa-1(21),7(22),8,10,12,14,17,19-octaen-19-yl]benzonitrile, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategies toward improving the brain penetration of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6IYS
 
 | |
2QZL
 
 | | Crystal Structure of human Beta Secretase complexed with IXS | | Descriptor: | Beta-secretase 1, N-[(1S)-1-benzyl-2-{[(1S)-2-(isobutylamino)-1-methyl-2-oxoethyl]amino}ethyl]-N'-[(1R)-1-(4-fluorophenyl)ethyl]-5-[methyl(methylsulfonyl)amino]isophthalamide | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BACE-1 inhibition by a series of psi[CH2NH] reduced amide isosteres
Bioorg.Med.Chem.Lett., 16, 2006
|
|
