3WV8
 
 | | ATP-bound HcgE from Methanothermobacter marburgensis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hmd co-occurring protein HcgE, SULFATE ION | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
2DC5
 
 | | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution | | Descriptor: | Glutathione S-transferase, mu 7 | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-28 | | Release date: | 2006-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution
To be Published
|
|
1MFK
 
 | | Structure of Prokaryotic SECIS mRNA Hairpin | | Descriptor: | 5'-R(P*GP*GP*CP*GP*GP*UP*UP*GP*CP*AP*GP*GP*UP*CP*UP*GP*CP*AP*CP*CP*GP*CP*C)-3' | | Authors: | Fourmy, D, Guittet, E, Yoshizawa, S. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Prokaryotic SECIS mRNA Hairpin and its Interaction with Elongation Factor SELB
J.Mol.Biol., 324, 2002
|
|
2DJN
 
 | | The solution structure of the homeobox domain of human Homeobox protein DLX-5 | | Descriptor: | Homeobox protein DLX-5 | | Authors: | Sasagawa, A, Ohnishi, S, Tochio, N, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-05 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeobox domain of human Homeobox protein DLX-5
To be Published
|
|
1U6I
 
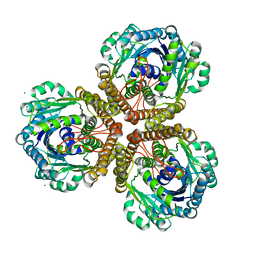 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2DMN
 
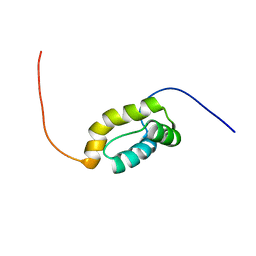 | | The solution structure of the homeobox domain of human homeobox protein TGIF2LX | | Descriptor: | Homeobox protein TGIF2LX | | Authors: | Tochio, N, Ohnishi, S, Sasagawa, A, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-22 | | Release date: | 2006-10-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeobox domain of human homeobox protein TGIF2LX
To be Published
|
|
1U6K
 
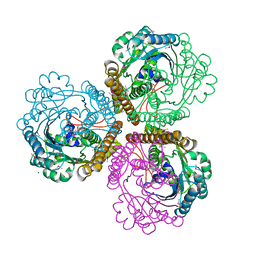 | | TLS refinement of the structure of Se-methionine labelled Coenzyme f420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) from Methanopyrus kandleri | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2DST
 
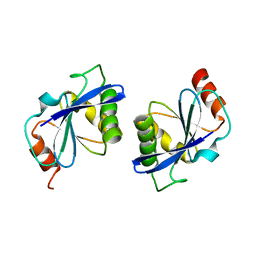 | | Crystal Structure Analysis of TT1977 | | Descriptor: | Hypothetical protein TTHA1544 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Shirouzu, M, Chen, L, Liu, Z.J, Wang, B.C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-06 | | Release date: | 2007-01-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the minimized alpha/beta-hydrolase fold protein from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1QNZ
 
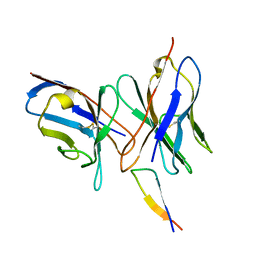 | | NMR structure of the 0.5b anti-HIV antibody complex with the gp120 V3 peptide | | Descriptor: | 0.5B ANTIBODY (HEAVY CHAIN), 0.5B ANTIBODY (LIGHT CHAIN), GP120 | | Authors: | Tugarinov, V, Zvi, A, Levy, R, Hayek, Y, Matsushita, S, Anglister, J. | | Deposit date: | 1999-10-26 | | Release date: | 2000-06-03 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Anti-Gp120 Antibody Complex with a V3 Peptide Reveals a Surface Important for Co-Receptor Binding
Structure, 8, 2000
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
2CZE
 
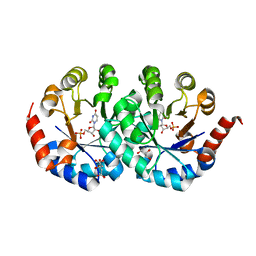 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Arai, R, Ito, K, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP
To be Published
|
|
2CY9
 
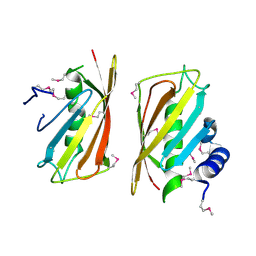 | | Crystal structure of thioesterase superfamily member2 from Mus musculus | | Descriptor: | Thioesterase superfamily member 2 | | Authors: | Hosaka, T, Murayama, K, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of thioesterase superfamily member2 from Mus musculus
To be Published
|
|
2CWE
 
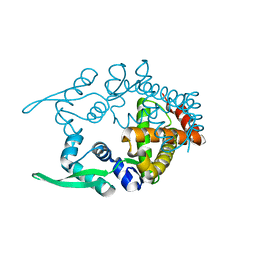 | | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical transcription regulator protein, PH1932 | | Authors: | Arai, R, Kishishita, S, Kukimoto-Niino, M, Wang, H, Sugawara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3
To be Published
|
|
3ACT
 
 | |
2LAA
 
 | |
2LAB
 
 | |
1QV9
 
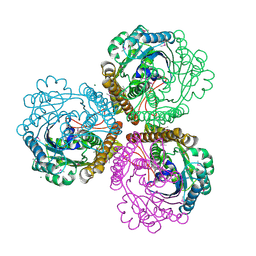 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
3VJ7
 
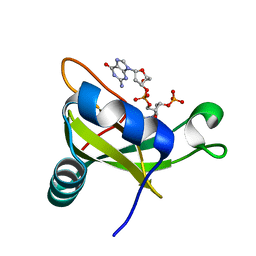 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 R33Q mutant | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
2DUM
 
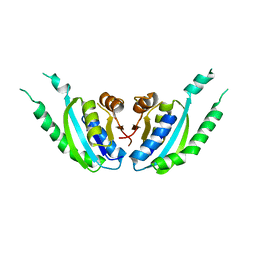 | | Crystal structure of hypothetical protein, PH0823 | | Descriptor: | Hypothetical protein PH0823 | | Authors: | Hosaka, T, Kishishita, S, Murayama, K, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-24 | | Release date: | 2007-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of hypothetical protein, PH0823
To be Published
|
|
1RHC
 
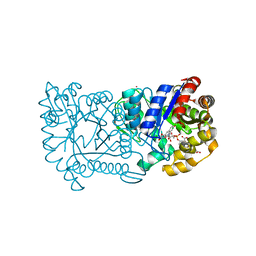 | | F420-dependent secondary alcohol dehydrogenase in complex with an F420-acetone adduct | | Descriptor: | ACETONE, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Berk, H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coenzyme binding in f(420)-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family.
Structure, 12, 2004
|
|
3AQV
 
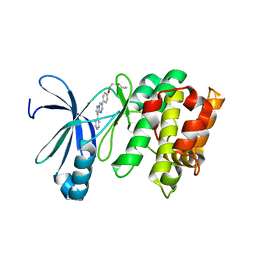 | | Human AMP-activated protein kinase alpha 2 subunit kinase domain (T172D) complexed with compound C | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine | | Authors: | Handa, N, Takagi, T, Saijo, S, Kishishita, S, Toyama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for compound C inhibition of the human AMP-activated protein kinase alpha 2 subunit kinase domain
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1UGP
 
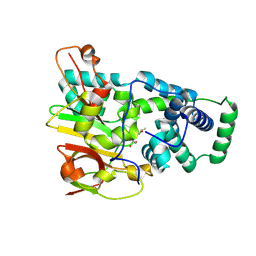 | | Crystal structure of Co-type nitrile hydratase complexed with n-butyric acid | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGS
 
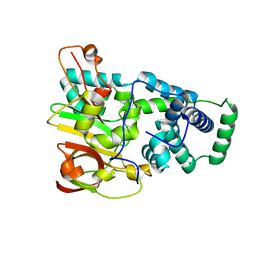 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
2DTC
 
 | | Crystal structure of MS0666 | | Descriptor: | RAL GUANINE NUCLEOTIDE EXCHANGE FACTOR RALGPS1A | | Authors: | Wang, H, Kishishita, S, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-12 | | Release date: | 2007-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MS0666
To be Published
|
|
