1J3T
 
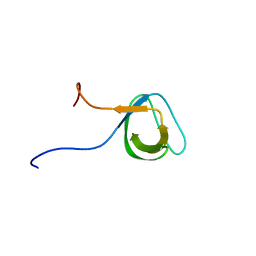 | | Solution structure of the second SH3 domain of human intersectin 2 (KIAA1256) | | Descriptor: | Intersectin 2 | | Authors: | Nameki, N, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-13 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH3 domain of human intersectin 2 (KIAA1256)
To be Published
|
|
6M5O
 
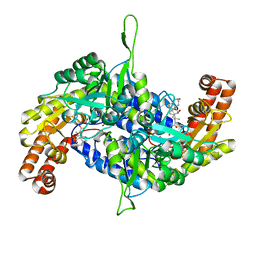 | | Co-crystal structure of human serine hydroxymethyltransferase 2 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, Serine hydroxymethyltransferase, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30000663 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
2E88
 
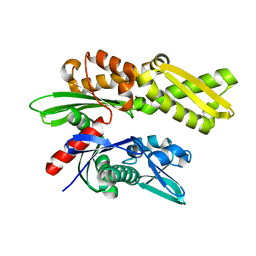 | | Crystal structure of the human Hsp70 ATPase domain in the apo form | | Descriptor: | Heat shock 70kDa protein 1A, ZINC ION | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1J2A
 
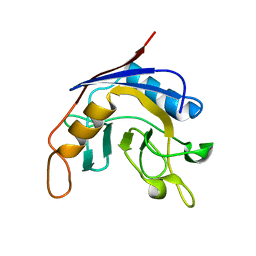 | | Structure of E. coli cyclophilin B K163T mutant | | Descriptor: | cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2002-12-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1V48
 
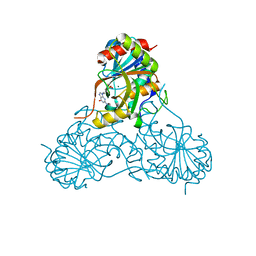 | | Calf spleen purine nucleoside phosphorylase (PNP) binary complex with 9-(5,5-difluoro-5-phosphonopenthyl)guanine | | Descriptor: | 9-(5,5-DIFLUORO-5-PHOSPHONOPENTYL)GUANINE, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Luic, M, Koellner, G, Yokomatsu, T, Shibuya, S, Bzowska, A. | | Deposit date: | 2003-11-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calf spleen purine-nucleoside phosphorylase: crystal structure of the binary complex with a potent multisubstrate analogue inhibitor.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
4P7W
 
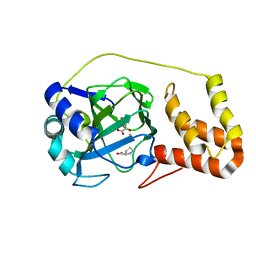 | | L-proline-bound L-proline cis-4-hydroxylase | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, L-proline cis-4-hydroxylase, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
6S1E
 
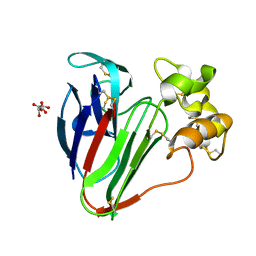 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
4UBS
 
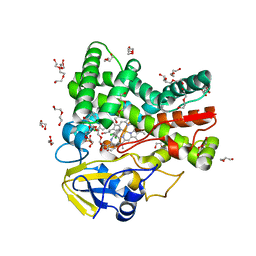 | | The crystal structure of cytochrome P450 105D7 from Streptomyces avermitilis in complex with Diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Xu, L.H, Ikeda, H, Arakawa, T, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6P86
 
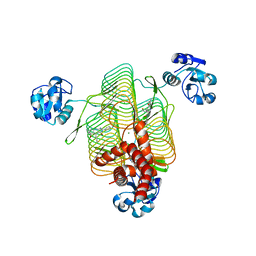 | | E.coli LpxD in complex with compound 4.1 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-[(4-ethylphenyl)methyl]-2-[(6aR)-6-oxo-3-(pyrrolidine-1-carbonyl)-6,6a,7,8,9,10-hexahydro-5H-pyrido[1,2-a]quinoxalin-5-yl]acetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P88
 
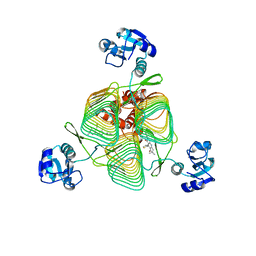 | | E.coli LpxD in complex with compound 6 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-{3-[(furan-2-carbonyl)amino]phenyl}-2,3-dihydro-1,4-benzodioxine-6-carboxamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
1J0G
 
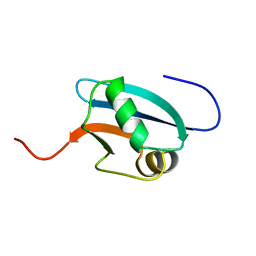 | | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold | | Descriptor: | Hypothetical Protein 1810045K17 | | Authors: | Zhao, C, Kigawa, T, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold
To be Published
|
|
6P85
 
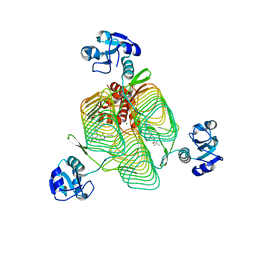 | | E.coli LpxD in complex with compound 3 | | Descriptor: | 1-(2-ethoxyphenyl)-4-[(4-methylphenyl)sulfonyl]-1H-1,2,3-triazol-5-amine, 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P89
 
 | | E.coli LpxD in complex with compound 7 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(8-methyl[1]benzopyrano[4,3-c]pyrazol-1(4H)-yl)acetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P87
 
 | | E.coli LpxD in complex with compound 5 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-[(R)-(1H-indol-3-yl)(thiophen-2-yl)methyl]-2-(2-methoxyphenoxy)-N-methylacetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P8B
 
 | | E.coli LpxD in complex with peptide FITC-RJPXD33 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, FITC-RJPXD33, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6FRN
 
 | | Structure of F420H2 oxidase (FprA) co-crystallized with 10mM Tb-Xo4 and calcium chloride | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, F420H2 oxidase (FprA), ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Wagner, T, Shima, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6P84
 
 | | E.coli LpxD in complex with compound 2o | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
4YT4
 
 | |
6P83
 
 | | E.coli LpxD in complex with compound 1o | | Descriptor: | 4-(2-chlorophenyl)-3-hydroxy-7,7-dimethyl-2-phenyl-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P8A
 
 | | E.coli LpxD in complex with compound 8.1 | | Descriptor: | 2-[4,6-dimethyl-3-(1H-pyrrol-1-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl]-N-[3-(morpholin-4-yl)propyl]acetamide, 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
4YT8
 
 | | SeMet-labelled HmdII from Methanocaldococcus jannaschii | | Descriptor: | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
4YT5
 
 | | HmdII from Methanocaldococcus jannaschii with bound methylene-tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
4WH3
 
 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
6FRM
 
 | | Crystal Structure of coenzyme F420H2 oxidase (FprA) co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Coenzyme F420H2 oxidase (FprA), FE (III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Wagner, T, Di Pietro, S, Shima, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
