1IT8
 
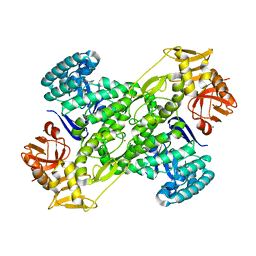 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
5B83
 
 | |
5I20
 
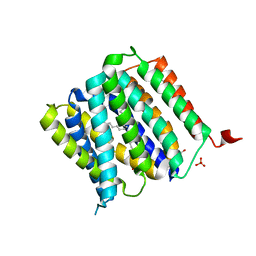 | | Crystal structure of protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | Authors: | Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
8IC7
 
 | | exo-beta-D-arabinofuranosidase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with beta-D-arabinofuranose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC6
 
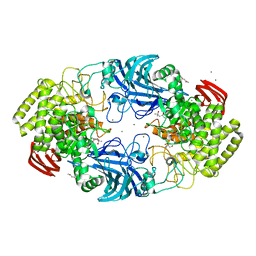 | | exo-beta-D-arabinanase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
1J2B
 
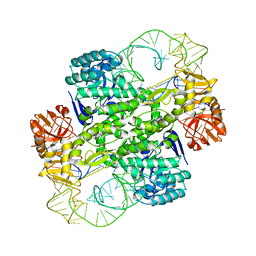 | | Crystal Structure Of Archaeosine tRNA-Guanine Transglycosylase Complexed With lambda-form tRNA(Val) | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Nameki, N, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-29 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternative Tertiary Structure of tRNA for Recognition by a Posttranscriptional Modification Enzyme
Cell(Cambridge,Mass.), 113, 2003
|
|
8J5H
 
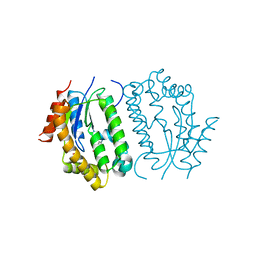 | | Crystal structure of kinase AbmG in complex with 4'-hydroxy-4'-thiocytidine | | Descriptor: | 4-azanyl-1-[(2~{R},3~{R},4~{S},5~{S})-5-(hydroxymethyl)-3,4,5-tris(oxidanyl)thiolan-2-yl]pyrimidin-2-one, Deoxyadenosine/deoxycytidine kinase, NITRATE ION | | Authors: | Ushimaru, R, Mori, T, Liu, H.-w, Abe, I. | | Deposit date: | 2023-04-22 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of kinase AbmG in complex with 4'-hydroxy-4'-thiocytidine
To Be Published
|
|
8J5G
 
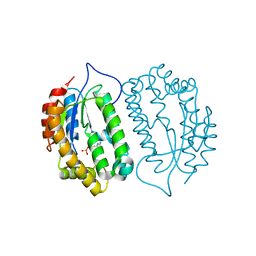 | | Crystal structure of kinase AbmG in complex with 4'-hydroxy-4'-thiocytidine 5'-diphosphate | | Descriptor: | Deoxyadenosine/deoxycytidine kinase, [(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-2-(hydroxymethyl)-3,4-bis(oxidanyl)thiolan-2-yl] phosphono hydrogen phosphate | | Authors: | Ushimaru, R, Mori, T, Liu, H.-w, Abe, I. | | Deposit date: | 2023-04-22 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of kinase AbmG in complex with 4'-hydroxy-4'-thiocytidine 5'-diphosphate
To Be Published
|
|
8J5F
 
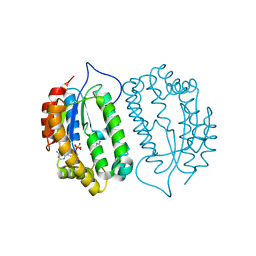 | |
8J5E
 
 | | Crystal structure of kinase AbmG in complex with 4'-thiocytidine | | Descriptor: | 4-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)thiolan-2-yl]pyrimidin-2-one, Deoxyadenosine/deoxycytidine kinase | | Authors: | Ushimaru, R, Mori, T, Liu, H.-w, Abe, I. | | Deposit date: | 2023-04-22 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of kinase AbmG in complex with 4'-thiocytidine
To Be Published
|
|
7XSG
 
 | | Crystal structure of ClAgl29B | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSF
 
 | | Crystal structure of ClAgl29A | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSH
 
 | | Crystal structure of ClAgl29B bound with L-glucose | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
8JQQ
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus C347T mutant | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQP
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQO
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with imidazole | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1A
 
 | | Photoswitchable fluorescent protein Gamillus, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
7E4L
 
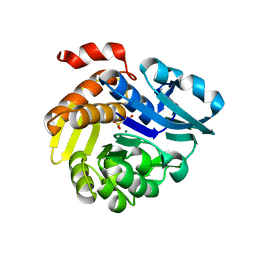 | |
6IE8
 
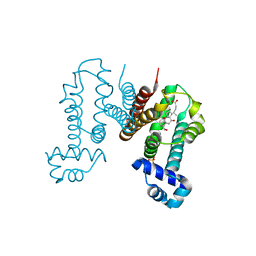 | | RamR in complex with cholic acid | | Descriptor: | CHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
6JXF
 
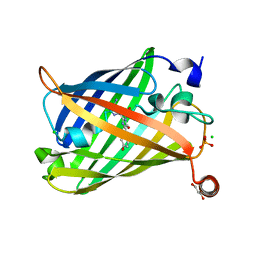 | | Photoswitchable fluorescent protein Gamillus, off-state (pH7.0) | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
8IAX
 
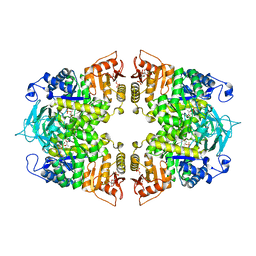 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with phosphoenolpyruvate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
8IAT
 
 | |
8IAS
 
 | |
