4URL
 
 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | 分子名称: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URN
 
 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | 分子名称: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-07-01 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4U9L
 
 | | Structure of a membrane protein | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MAGNESIUM ION, Magnesium transporter MgtE | | 著者 | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | 登録日 | 2014-08-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
4U9N
 
 | | Structure of a membrane protein | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MANGANESE (II) ION, Magnesium transporter MgtE, ... | | 著者 | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | 登録日 | 2014-08-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
8JHK
 
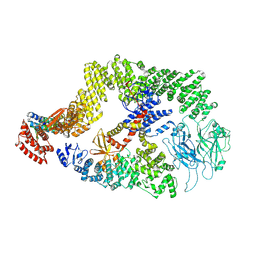 | | Cryo-EM structure of the DOCK5/ELMO1 complex, focused on one protomer | | 分子名称: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1 | | 著者 | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (4.76 Å) | | 主引用文献 | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
1UFA
 
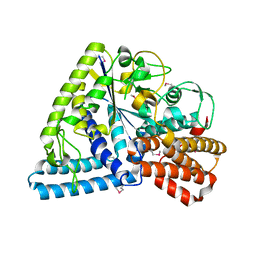 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | 分子名称: | TT1467 protein | | 著者 | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-05-28 | | 公開日 | 2003-11-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
1V6Q
 
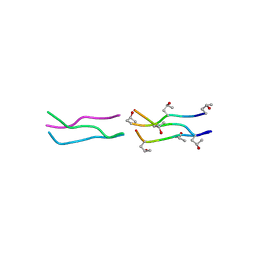 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.3 A | | 分子名称: | Collagen like peptide | | 著者 | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | 登録日 | 2003-12-03 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
6LNG
 
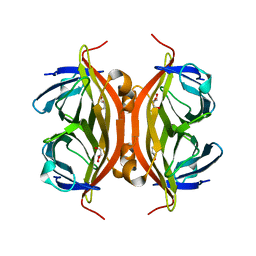 | | Rapid crystallization of streptavidin using charged peptides | | 分子名称: | GLYCEROL, Streptavidin | | 著者 | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | 登録日 | 2019-12-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8000015 Å) | | 主引用文献 | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
5B1O
 
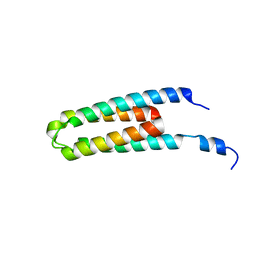 | | DHp domain structure of EnvZ P248A mutant | | 分子名称: | Osmolarity sensor protein EnvZ | | 著者 | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
5B1N
 
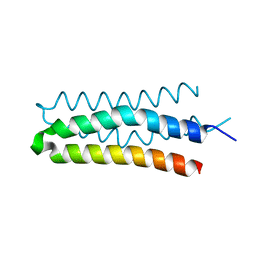 | | DHp domain structure of EnvZ from Escherichia coli | | 分子名称: | Osmolarity sensor protein EnvZ | | 著者 | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
1ULH
 
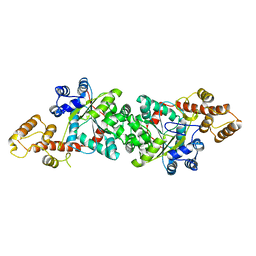 | | A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase | | 分子名称: | Tryptophanyl-tRNA synthetase | | 著者 | Kise, Y, Sengoku, T, Ishii, R, Yokoyama, S, Park, S.G, Lee, S.W, Kim, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-09-12 | | 公開日 | 2004-02-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase
Nat.Struct.Mol.Biol., 11, 2004
|
|
4XJ6
 
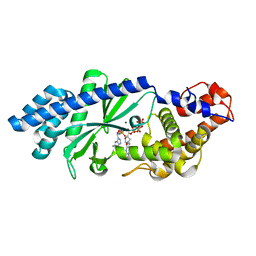 | | Crystal structure of Escherichia coli DncV 3'-deoxy GTP bound form | | 分子名称: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, VC0179-like protein | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ3
 
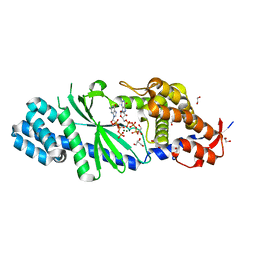 | | Crystal structure of Vibrio cholerae DncV GTP bound form | | 分子名称: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
1V2X
 
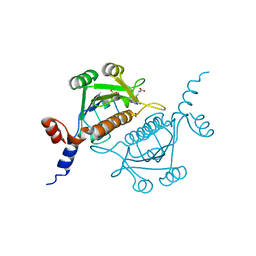 | | TrmH | | 分子名称: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (Gm18) methyltransferase | | 著者 | Nureki, O, Watanabe, K, Fukai, S, Ishii, R, Endo, Y, Hori, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-10-17 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Deep Knot Structure for Construction of Active Site and Cofactor Binding Site of tRNA Modification Enzyme
STRUCTURE, 12, 2004
|
|
4XJ1
 
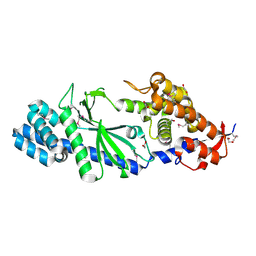 | | Crystal structure of Vibrio cholerae DncV apo form | | 分子名称: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ4
 
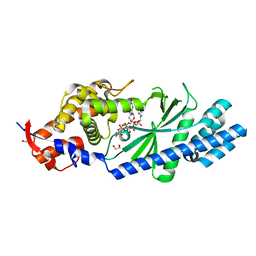 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy ATP bound form | | 分子名称: | 1,2-ETHANEDIOL, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ5
 
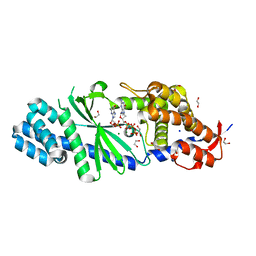 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy GTP bound form | | 分子名称: | 1,2-ETHANEDIOL, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.552 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4PDN
 
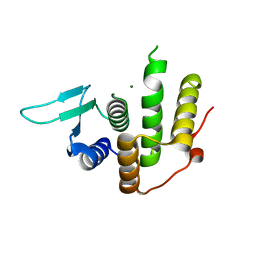 | | Crystal structure of E. coli YfcM | | 分子名称: | MAGNESIUM ION, Uncharacterized protein | | 著者 | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2014-04-19 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.448 Å) | | 主引用文献 | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation.
Nucleic Acids Res., 42, 2014
|
|
1B90
 
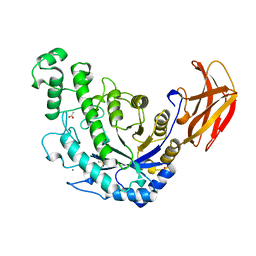 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | 分子名称: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | 著者 | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | 登録日 | 1999-03-06 | | 公開日 | 1999-03-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
3WUH
 
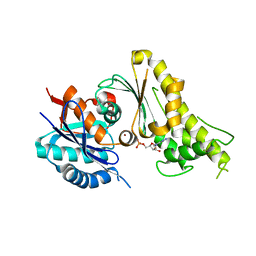 | | Qri7 and AMP complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, ZINC ION, tRNA N6-adenosine threonylcarbamoyltransferase, ... | | 著者 | Tominaga, T, Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2014-04-24 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.937 Å) | | 主引用文献 | Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
3WTR
 
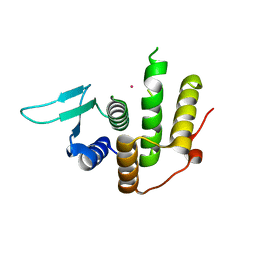 | | Crystal structure of E. coli YfcM bound to Co(II) | | 分子名称: | COBALT (II) ION, Uncharacterized protein | | 著者 | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2014-04-19 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation
Nucleic Acids Res., 42, 2014
|
|
4YZI
 
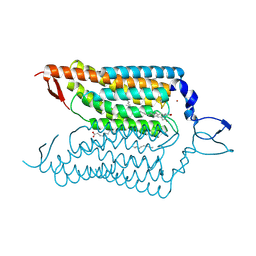 | | Crystal structure of blue-shifted channelrhodopsin mutant (T198G/G202A) | | 分子名称: | OLEIC ACID, RETINAL, Sensory opsin A,Archaeal-type opsin 2, ... | | 著者 | Kato, H.E, Kamiya, M, Ishitani, R, Hayashi, S, Nureki, O. | | 登録日 | 2015-03-25 | | 公開日 | 2015-05-27 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Atomistic design of microbial opsin-based blue-shifted optogenetics tools.
Nat Commun, 6, 2015
|
|
6LX6
 
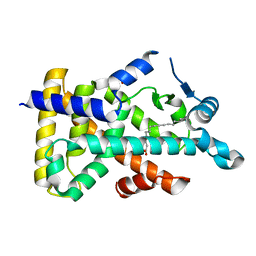 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | 分子名称: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXB
 
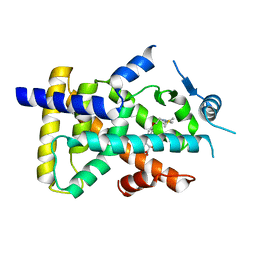 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | 分子名称: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXA
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | 分子名称: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
