1IVJ
 
 | | Crystal Structure of Rat Hemeoxygenase-1 in Complex with Heme and Azide. | | Descriptor: | AZIDE ION, Hemeoxygenase-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-03-18 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rat Heme Oxygenase-1 in Complex with Heme Bound to Azide. IMPLICATION FOR REGIOSPECIFIC HYDROXYLATION OF HEME AT THE alpha -MESO CARBON
J.Biol.Chem., 277, 2002
|
|
7EGO
 
 | | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527 | | Descriptor: | 3-[methyl-(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]propanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Takabayashi, M, Yokota, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Sugiyama, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527
To Be Published
|
|
5BUN
 
 | | Crystal structure of an antigenic outer membrane protein ST50 from Salmonella Typhi | | Descriptor: | Outer membrane protein, octyl beta-D-glucopyranoside | | Authors: | Yoshimura, M, Chuankhayan, P, Lin, C.C, Chen, N.C, Yang, M.C, Fun, H.K. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of an antigenic outer-membrane protein from Salmonella Typhi suggests a potential antigenic loop and an efflux mechanism.
Sci Rep, 5, 2015
|
|
1IX4
 
 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Carbon Monoxide | | Descriptor: | CARBON MONOXIDE, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
6L0R
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Acetylserine | | Descriptor: | (2S)-3-acetyloxy-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0S
 
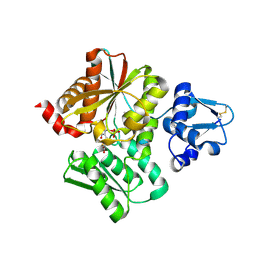 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with L-Cysteine | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
4YK7
 
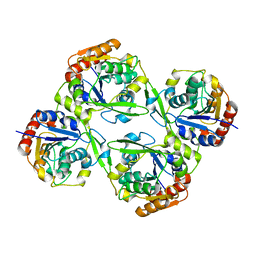 | |
2ZMI
 
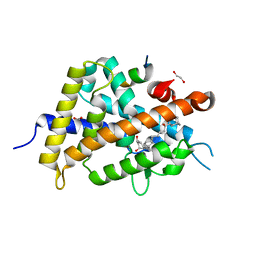 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]pent-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2ZMJ
 
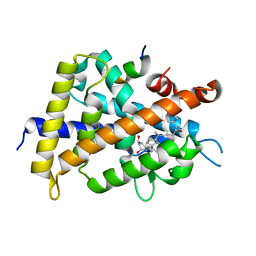 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
1G4F
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
1G4G
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
6E2H
 
 | |
2ZMH
 
 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1R,2E,4R)-4-hydroxy-1-methyl-4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]but-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
3CA9
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-02-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8GZC
 
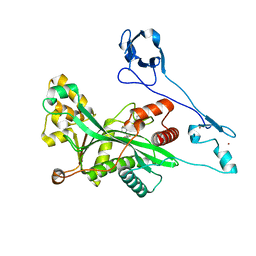 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
8HVA
 
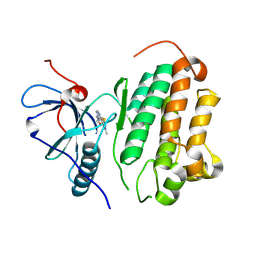 | |
8IHL
 
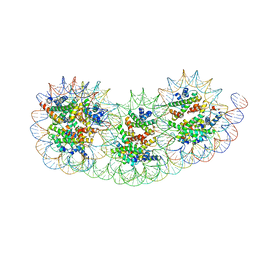 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
7VI0
 
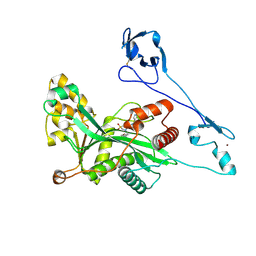 | | Crystal structure of EP300 HAT domain in complex with compound 11 | | Descriptor: | (4S)-N-(3H-indazol-4-yl)-3-[1-(4-methoxyphenyl)cyclopentyl]carbonyl-1,1-bis(oxidanylidene)-1,3-thiazolidine-4-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
7VHZ
 
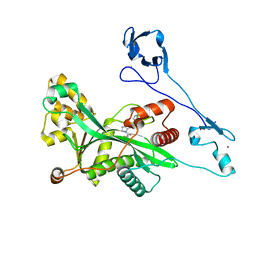 | | Crystal structure of EP300 HAT domain in complex with compound 7 | | Descriptor: | (2R)-N-(2H-indazol-4-yl)-1-[1-(4-methoxyphenyl)cyclopentyl]carbonyl-pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
7VHY
 
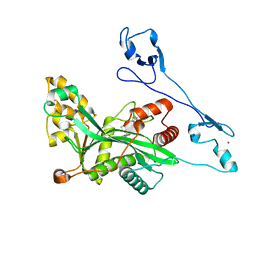 | | Crystal structure of EP300 HAT domain in complex with compound (+)-3 | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(6R)-6-(1H-indazol-4-ylmethyl)-1,4-oxazepan-4-yl]-[1-(4-methoxyphenyl)cyclopentyl]methanone | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
2E8A
 
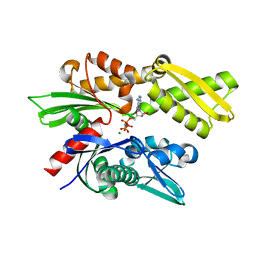 | | Crystal structure of the human Hsp70 ATPase domain in complex with AMP-PNP | | Descriptor: | Heat shock 70kDa protein 1A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2Z6E
 
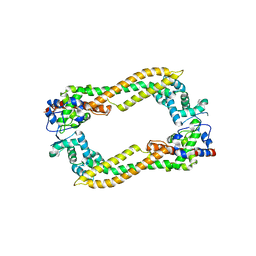 | | Crystal Structure of Human DAAM1 FH2 | | Descriptor: | Disheveled-associated activator of morphogenesis 1 | | Authors: | Yamashita, M, Higashi, T, Sato, Y, Shirakawa, R, Kita, T, Horiuchi, H, Fukai, S, Nureki, O. | | Deposit date: | 2007-07-31 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human DAAM1 formin homology 2 domain
Genes Cells, 12, 2007
|
|
2E88
 
 | | Crystal structure of the human Hsp70 ATPase domain in the apo form | | Descriptor: | Heat shock 70kDa protein 1A, ZINC ION | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6E29
 
 | |
2QKS
 
 | |
