2DNU
 
 | | Solution structure of RSGI RUH-061, a SH3 domain from human | | Descriptor: | SH3 multiple domains 1 | | Authors: | Ohashi, W, Hirota, H, Nagashima, T, Kurosaki, C, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-061, a SH3 domain from human
to be published
|
|
6M04
 
 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
1UFN
 
 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
5KNC
 
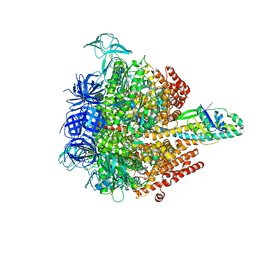 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
4XD7
 
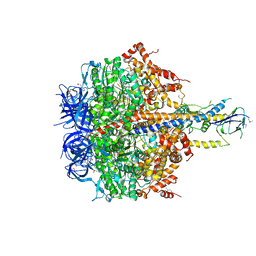 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
5KNB
 
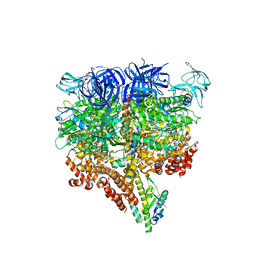 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
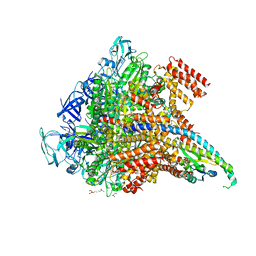 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
1UHT
 
 | | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein | | Descriptor: | expressed protein | | Authors: | Tomizawa, T, Inoue, M, Koshiba, S, Hayashi, F, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Shinozaki, K, Seki, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein
To be Published
|
|
6EII
 
 | | The crystal structure of CK2alpha in complex with compound 18 | | Descriptor: | (3-chloranyl-4-phenyl-phenyl)methyl-(3-phenylpropyl)azanium, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1HMK
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
6EHK
 
 | | The crystal structure of CK2alpha in complex with CAM4712 and compound 37 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[3,5-bis(chloranyl)-4-(2-ethylphenyl)phenyl]methyl]ethanamine, 2-hydroxy-5-methylbenzoic acid, ACETATE ION, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6EHU
 
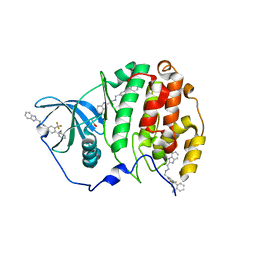 | | The crystal structure of CK2alpha in complex with compound 32 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[4-(2-ethylphenyl)-3-(trifluoromethyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
7PCE
 
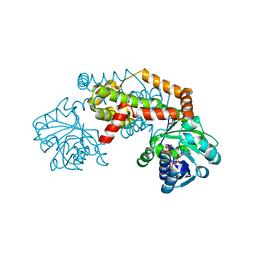 | | BurG (apo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketol-acid reductoisomerase, PHOSPHATE ION | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCC
 
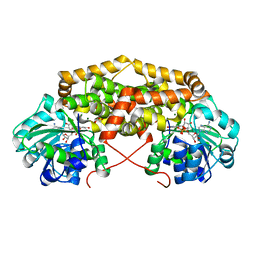 | | BurG in complex with Mg2+ and NAD+ (holo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCG
 
 | | BurG (holo) in complex with cyclopropane-1,1-dicarboxylate (7): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCM
 
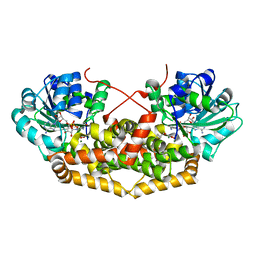 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCO
 
 | | BurG E232Q mutant (holo) in complex with 2R,3R-2,3-dihydroxy-6-methyl-heptanoate (12): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R,3R)-6-methyl-2,3-bis(oxidanyl)heptanoic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCI
 
 | | BurG (holo) in complex with hydroxypyruvate-enol (8): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-2,3-bis(oxidanyl)prop-2-enoic acid, IMIDAZOLE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCT
 
 | | BurG E232Q mutant (holo) in complex with enol-oxalacetate (15): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCL
 
 | | BurG (holo) in complex with 2-hydroxy-2-(hydroxy(isopropyl)amino)acetate (11): Biosynthesis of cyclopropanolrings in bacterial toxins | | Descriptor: | (2S)-2-oxidanyl-2-[oxidanyl(propan-2-yl)amino]ethanoic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCN
 
 | | BurG (holo) in complex with gonyenediol (14), trigonic acid (6) and DMS: Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R)-2-oxidanyl-2-(1-oxidanylcyclopropyl)ethanoic acid, (METHYLSULFANYL)METHANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
3KB1
 
 | | Crystal Structure of the Nucleotide-binding protein AF_226 in complex with ADP from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR157 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleotide-binding protein, ZINC ION | | Authors: | Forouhar, F, Lew, S, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR157
To be Published
|
|
3KAT
 
 | | Crystal Structure of the CARD domain of the human NLRP1 protein, Northeast Structural Genomics Consortium Target HR3486E | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR3486E
To be Published
|
|
1QVR
 
 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
