8FR5
 
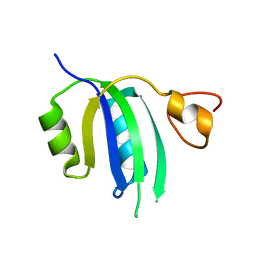 | | Crystal structure of the Human Smacovirus 1 Rep domain | | 分子名称: | MANGANESE (II) ION, Rep, SODIUM ION | | 著者 | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | 登録日 | 2023-01-06 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6Q1M
 
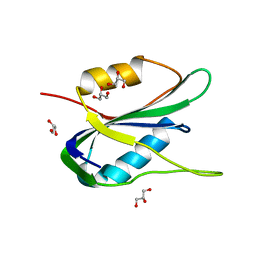 | | Crystal structure of the wheat dwarf virus Rep domain | | 分子名称: | GLYCEROL, Replication-associated protein | | 著者 | Litzau, L.A, Everett, B.A, Evans III, R.L, Shi, K, Tompkins, K, Gordon, W.R. | | 登録日 | 2019-08-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystal structure of the Wheat dwarf virus Rep domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | 分子名称: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | 著者 | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | 登録日 | 2022-08-09 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
6VOY
 
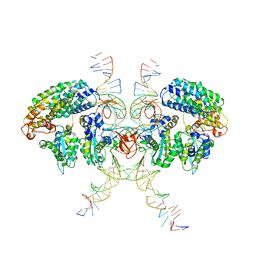 | | Cryo-EM structure of HTLV-1 instasome | | 分子名称: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | 著者 | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | 登録日 | 2020-02-01 | | 公開日 | 2020-07-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
6O44
 
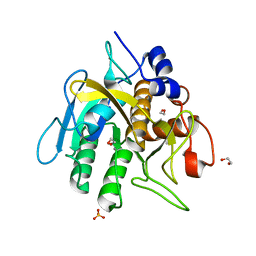 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | 著者 | Tang, H, Shi, K, Aihara, H. | | 登録日 | 2019-02-28 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
6WDZ
 
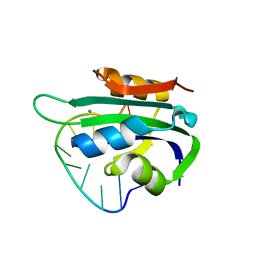 | | Porcine circovirus 2 Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | 分子名称: | 1,2-ETHANEDIOL, ATP-dependent helicase Rep, DNA (5'-D(*TP*AP*GP*TP*AP*TP*TP*AP*CP*C)-3'), ... | | 著者 | Litzau, L.A, Tompkins, K, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | 登録日 | 2020-04-01 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
6WE0
 
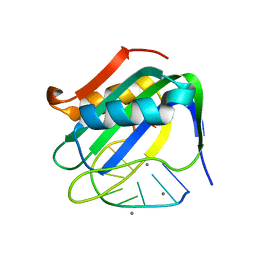 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | 分子名称: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*AP*CP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | 著者 | Tompkins, K, Litzau, L.A, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | 登録日 | 2020-04-01 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
8WOY
 
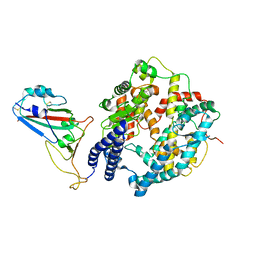 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with rabbit ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | 著者 | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | 登録日 | 2023-10-08 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOZ
 
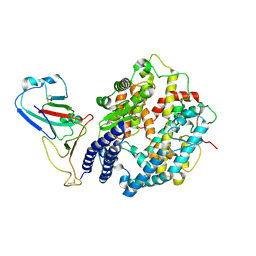 | | Cryo-EM structure of SARS-CoV RBD in complex with rabbit ACE2 | | 分子名称: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | 著者 | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | 登録日 | 2023-10-08 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOX
 
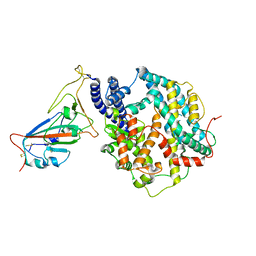 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with rabbit ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | 著者 | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | 登録日 | 2023-10-08 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8EJO
 
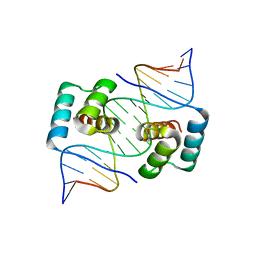 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
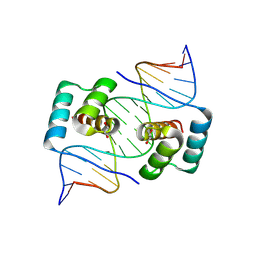 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EUO
 
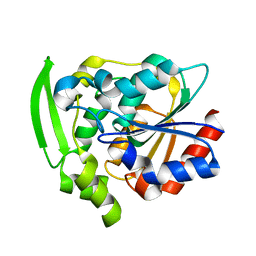 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | 分子名称: | (S)-hydroxynitrile lyase | | 著者 | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | 登録日 | 2022-10-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | to be published
To Be Published
|
|
6WGX
 
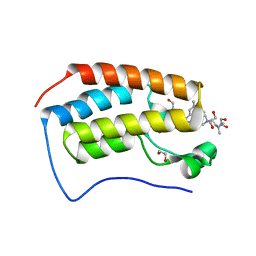 | | Cocrystal of BRD4(D1) with a selective inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-(1-{1-[2-(dimethylamino)ethyl]piperidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-5-yl)-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | 著者 | Johnson, J.A, Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2020-04-06 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | 著者 | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | 登録日 | 2021-08-19 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
7RWN
 
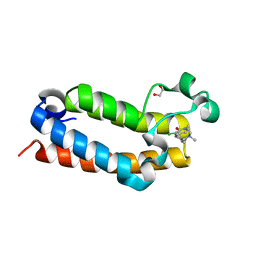 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWP
 
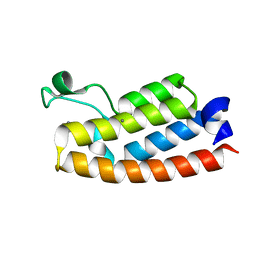 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | 分子名称: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
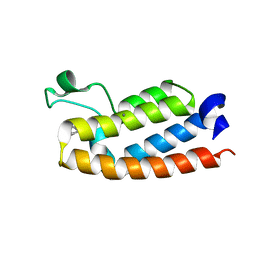 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | 分子名称: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
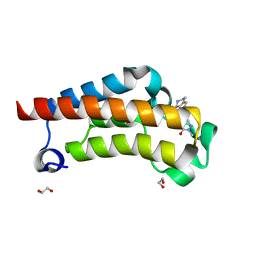 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
5IRS
 
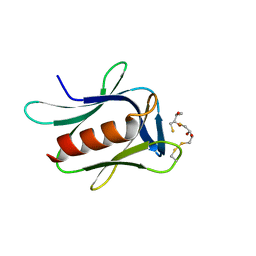 | | crystal structure of the proteasomal Rpn13 PRU-domain | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Proteasomal ubiquitin receptor ADRM1 | | 著者 | Chen, X, Shi, K, Walters, K, Aihara, H. | | 登録日 | 2016-03-14 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
8XLV
 
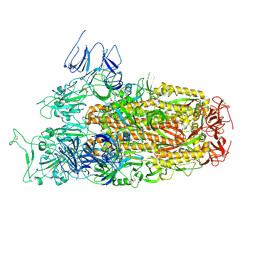 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-26 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XMT
 
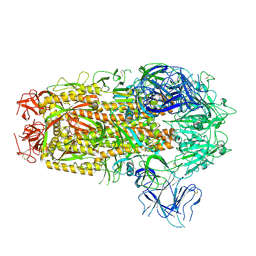 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-28 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN5
 
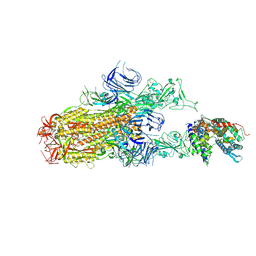 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-29 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNK
 
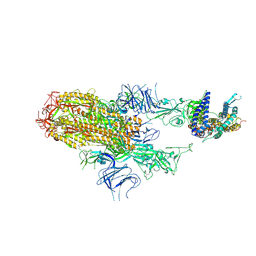 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-30 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XMG
 
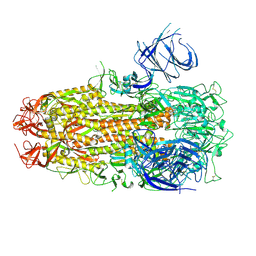 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-27 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
