8GQY
 
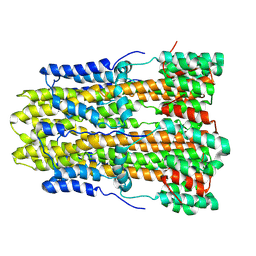 | | CryoEM structure of pentameric MotA from Aquifex aeolicus | | Descriptor: | Motility protein A | | Authors: | Nishikino, T, Takekawa, N, Kishikawa, J, Hirose, M, Onoe, S, Kato, T, Imada, K. | | Deposit date: | 2022-08-31 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of MotA, a flagellar stator protein, from hyperthermophile.
Biochem.Biophys.Res.Commun., 631, 2022
|
|
4D7Y
 
 | | Crystal structure of mouse C1QL1 globular domain | | Descriptor: | C1Q-RELATED FACTOR, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kakegawa, W, Mitakidis, N, Miura, E, Abe, M, Matsuda, K, Takeo, Y, Kohda, K, Motohashi, J, Takahashi, A, Nagao, S, Muramatsu, S, Watanabe, M, Sakimura, K, Aricescu, A.R, Yuzaki, M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Anterograde C1Ql1 Signaling is Required in Order to Determine and Maintain a Single-Winner Climbing Fiber in the Mouse Cerebellum
Neuron, 85, 2015
|
|
9IMA
 
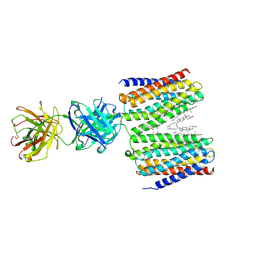 | | Cryo-EM structure for the GPRC5D complexed with Talquetamab Fab | | Descriptor: | CHOLESTEROL, G-protein coupled receptor family C group 5 member D, Talquetamab Fab (anti-GPRC5D) Heavy chain, ... | | Authors: | Jeong, J, Shin, J, Park, J, Cho, Y. | | Deposit date: | 2024-07-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural Basis for the Recognition of GPRC5D by Talquetamab, a Bispecific Antibody for Multiple Myeloma.
J.Mol.Biol., 436, 2024
|
|
4OM8
 
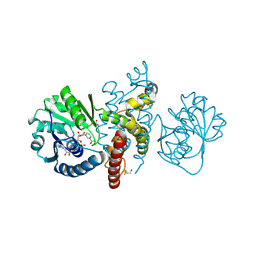 | | Crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) 5-dehydrogenase, an NAD+ dependent dismutase. | | Descriptor: | 3-hydroxybutyryl-coA dehydrogenase, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T, Ohnishi, K. | | Deposit date: | 2014-01-27 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Biochem.Biophys.Res.Commun., 456, 2015
|
|
4WCO
 
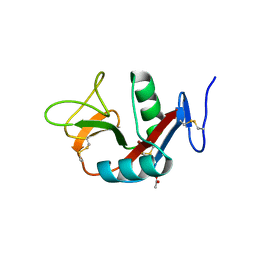 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
4YSV
 
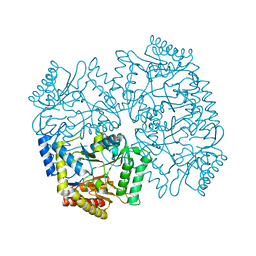 | |
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4TS6
 
 | |
8J3S
 
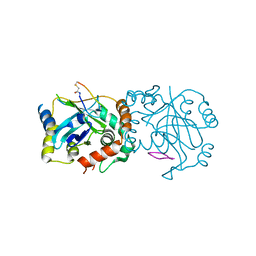 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
5EYT
 
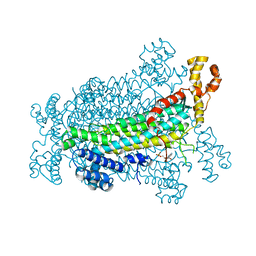 | | Crystal Structure of Adenylosuccinate Lyase from Schistosoma mansoni in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L, Nettleship, J, Owens, R, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3649 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
8GRJ
 
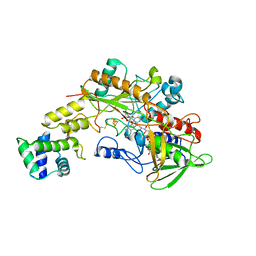 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Improvement of substrate specificity of the direct electron transfer type FAD-dependent glucose dehydrogenase catalytic subunit.
J.Biotechnol., 2024
|
|
4GRV
 
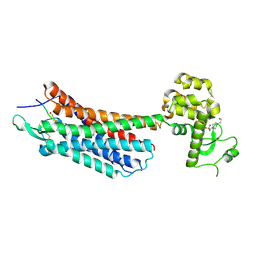 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
3CIY
 
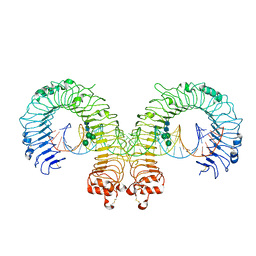 | | Mouse Toll-like receptor 3 ectodomain complexed with double-stranded RNA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
3CIG
 
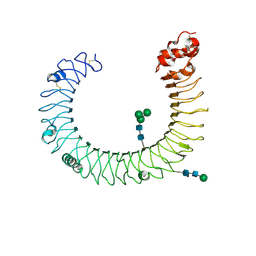 | | Crystal structure of mouse TLR3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
5GZ6
 
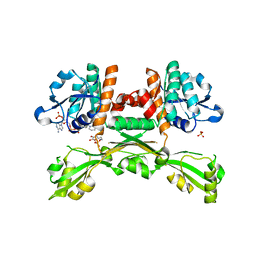 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | Descriptor: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
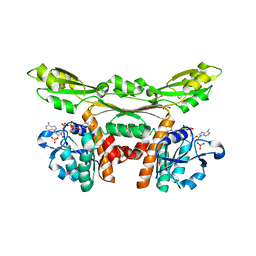 | | Structure of D-amino acid dehydrogenase in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ1
 
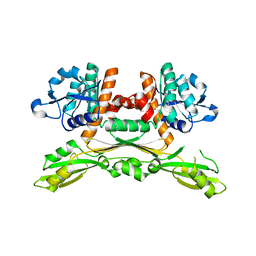 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5FB3
 
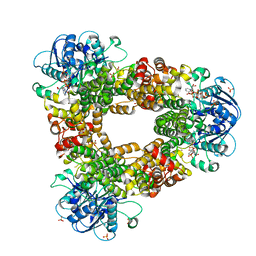 | | Structure of glycerophosphate dehydrogenase in complex with NADPH | | Descriptor: | Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE, ... | | Authors: | Sakuraba, H, Hayashi, J, Yamamoto, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique coenzyme binding mode of hyperthermophilic archaeal sn-glycerol-1-phosphate dehydrogenase from Pyrobaculum calidifontis
Proteins, 84, 2016
|
|
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BZB
 
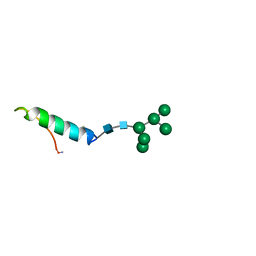 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
6K60
 
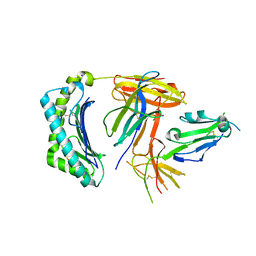 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
5O9M
 
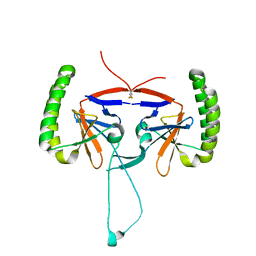 | | Crystal structure of human Histamine-Releasing Factor (HRF/TCTP)containing a disulphide-linked dimer | | Descriptor: | DI(HYDROXYETHYL)ETHER, Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
5O9L
 
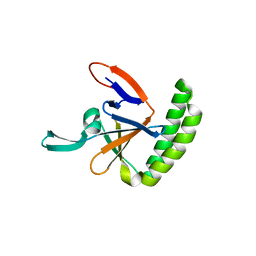 | | Crystal structure of human Histamine-Releasing Factor (HRF/TCTP) | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
5O9K
 
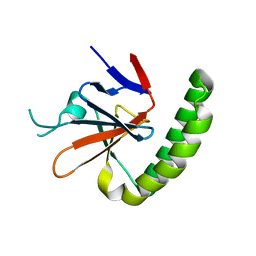 | | Crystal structure of Murine Histmaine-Releasing Factor (HRF/TCTP) | | Descriptor: | GLYCEROL, Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.014 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
