1KWK
 
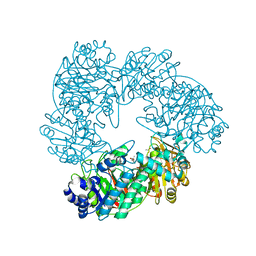 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
3I4D
 
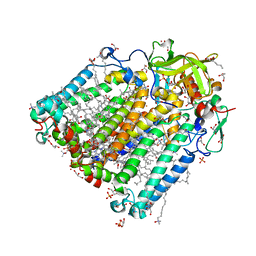 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
8GRT
 
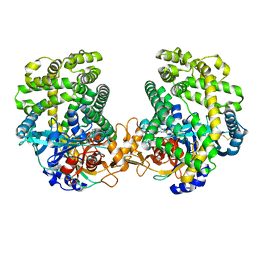 | | Small Dipeptide Analogues developed by Co-crystal Structure of Stenotrophomonas maltophilia Dipeptidyl Peptidase 7 | | Descriptor: | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID, Dipeptidyl-peptidase, TYROSINE | | Authors: | Yasumitsu, S, Koushi, H, Akihiro, N, Yoshiyuki, Y, Wataru, O, Mizuki, S, Saori, R, Nobutada, T, Anna, M, Keiko, H, Tsuda, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small Dipeptide Analogues Generated by Co-crystal Structure of Bacterial Dipeptidyl Peptidase 7 to Defeat Stenotrophomonas maltophilia
To Be Published
|
|
2ZVK
 
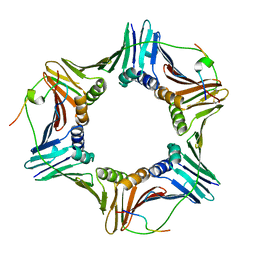 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1IWK
 
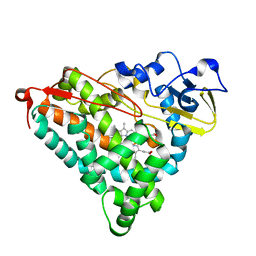 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(112K) Cytochrome P450cam | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
6D0D
 
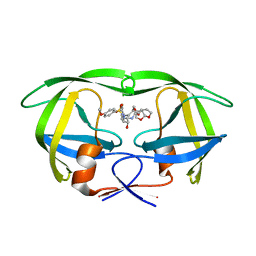 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-087-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
2ZVM
 
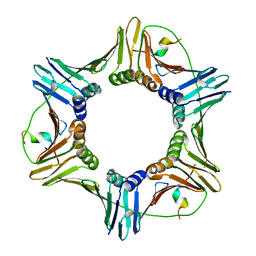 | | Crystal structure of PCNA in complex with DNA polymerase iota fragment | | Descriptor: | DNA polymerase iota, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
6D0E
 
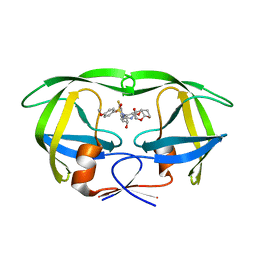 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-084-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
2ZVL
 
 | | Crystal structure of PCNA in complex with DNA polymerase kappa fragment | | Descriptor: | DNA polymerase kappa, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1IWI
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
1IWJ
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(109K) Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
1GEB
 
 | | X-RAY CRYSTAL STRUCTURE AND CATALYTIC PROPERTIES OF THR252ILE MUTANT OF CYTOCHROME P450CAM | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hishiki, T, Shimada, H, Nagano, S, Park, S.-Y, Ishimura, Y. | | Deposit date: | 2000-11-01 | | Release date: | 2000-11-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure and catalytic properties of Thr252Ile mutant of cytochrome P450cam: roles of Thr252 and water in the active center.
J.Biochem., 128, 2000
|
|
6KR6
 
 | | Crystal structure of Drosophila Piwi | | Descriptor: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | Authors: | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
3HJR
 
 | | Crystal structure of serine protease of Aeromonas sobria | | Descriptor: | CALCIUM ION, Extracellular serine protease | | Authors: | Utsunomiya, H, Tsuge, H, Kobayashi, H, Okamoto, K. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the kexin-like serine protease from Aeromonas sobria as a sepsis-causing factor
J.Biol.Chem., 284, 2009
|
|
2ZFD
 
 | | The crystal structure of plant specific calcium binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14 | | Descriptor: | ACETIC ACID, CALCIUM ION, Calcineurin B-like protein 2, ... | | Authors: | Akaboshi, M, Hashimoto, H, Ishida, H, Koizumi, N, Sato, M, Shimizu, T. | | Deposit date: | 2007-12-29 | | Release date: | 2008-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14
J.Mol.Biol., 377, 2008
|
|
8GUH
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Tris | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [4-[[[2-(hydroxymethyl)-1,3-bis(oxidanyl)propan-2-yl]amino]methyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Sphingobacterium multivorum serine palmitoyltransferase complexed with tris(hydroxymethyl)aminomethane.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8UH8
 
 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH9
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
1IXS
 
 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
8H20
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H21
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-alanine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H29
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-threonine | | Descriptor: | 1,2-ETHANEDIOL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1W
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
1IXR
 
 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
