1MKR
 
 | | Crystal Structure of a Mutant Variant of Cytochrome c Peroxidase (Plate like crystals) | | Descriptor: | Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
1ML2
 
 | | Crystal Structure of a Mutant Variant of Cytochrome c Peroxidase with Zn(II)-(20-oxo-Protoporphyrin IX) | | Descriptor: | 20-OXO-PROTOPORPHYRIN IX CONTAINING ZN(II), Cytochrome c Peroxidase | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
1MK8
 
 | | Crystal Structure of a Mutant Cytochrome c Peroxidase showing a Novel Trp-Tyr Covalent Cross-link | | Descriptor: | Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
1MKQ
 
 | | Crystal Structure of the Mutant Variant of Cytochrome c Peroxidase in the 'Open' Uncross-linked form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
4DVZ
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
4DVY
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
5H34
 
 | | Crystal structure of the C-terminal domain of methionyl-tRNA synthetase (MetRS-C) in Nanoarchaeum equitans | | Descriptor: | Methionine-tRNA ligase | | Authors: | Suzuki, H, Kaneko, A, Yamamoto, T, Nambo, M, Umehara, T, Yoshida, H, Park, S.Y, Tamura, K. | | Deposit date: | 2016-10-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Binding Properties of Split tRNA to the C-terminal Domain of Methionyl-tRNA Synthetase of Nanoarchaeum equitans.
J. Mol. Evol., 84, 2017
|
|
7C77
 
 | | Cryo-EM structure of mouse TLR3 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-05-23 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7C76
 
 | | Cryo-EM structure of human TLR3 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-05-23 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CYN
 
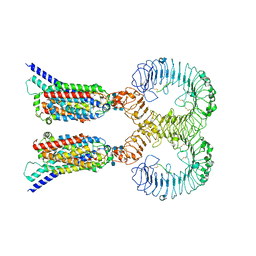 | | Cryo-EM structure of human TLR7 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1C7Y
 
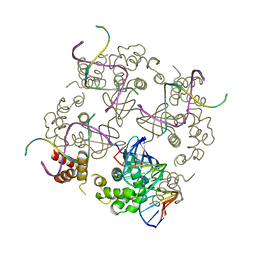 | | E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX | | Descriptor: | DNA (5'-D(P*DAP*DAP*DGP*DTP*DTP*DGP*DGP*DGP*DAP*DTP*DTP*DGP*DT)-3'), DNA (5'-D(P*DCP*DAP*DAP*DTP*DCP*DCP*DCP*DAP*DAP*DCP*DTP*DT)-3'), DNA (5'-D(P*DCP*DGP*DAP*DAP*DTP*DGP*DTP*DGP*DTP*DGP*DTP*DCP*DT)-3'), ... | | Authors: | Ariyoshi, M, Nishino, T, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1D8L
 
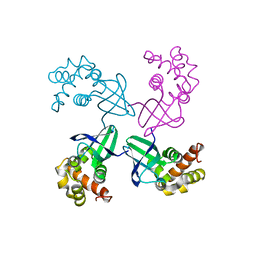 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
7CP7
 
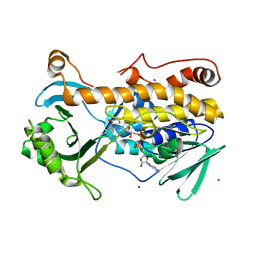 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
6UEP
 
 | |
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
6UER
 
 | |
6UEQ
 
 | | Structure of TBP bound to C-C mismatch containing TATA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*CP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), SULFATE ION, ... | | Authors: | Schumacher, M.A, Al-Hashimi, H. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
8GRT
 
 | | Small Dipeptide Analogues developed by Co-crystal Structure of Stenotrophomonas maltophilia Dipeptidyl Peptidase 7 | | Descriptor: | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID, Dipeptidyl-peptidase, TYROSINE | | Authors: | Yasumitsu, S, Koushi, H, Akihiro, N, Yoshiyuki, Y, Wataru, O, Mizuki, S, Saori, R, Nobutada, T, Anna, M, Keiko, H, Tsuda, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small Dipeptide Analogues Generated by Co-crystal Structure of Bacterial Dipeptidyl Peptidase 7 to Defeat Stenotrophomonas maltophilia
To Be Published
|
|
6QCU
 
 | |
6QD8
 
 | | EM structure of a EBOV-GP bound to 4M0368 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | rVSV-ZEBOV induces a polyclonal and convergent B cell response with potent Ebola virus-neutralizing antibodies
Nat.Med. (N.Y.), 2019
|
|
6QD7
 
 | | EM structure of a EBOV-GP bound to 3T0331 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV.
Nat. Med., 25, 2019
|
|
6KJG
 
 | | Crystal structure of PsoF | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
6KJI
 
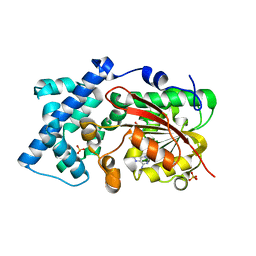 | | Crystal structure of PsoF with SAH | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
