5UZF
 
 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNA structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5UZD
 
 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A2-DNA structure | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*GP*AP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*TP*CP*GP*AP*TP*GP*C)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
6UH5
 
 | | Structural basis of COMPASS eCM recognition of the H2Bub nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
8THV
 
 | | FARFAR-NMR ensemble of HIV-1 TAR with apical loop capturing ground and excited conformational states | | Descriptor: | RNA (29-MER) | | Authors: | Roy, R, Geng, A, Shi, H, Merriman, D.K, Dethoff, E.A, Salmon, L, Al-Hashimi, H.M. | | Deposit date: | 2023-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.
J.Am.Chem.Soc., 145, 2023
|
|
8U3M
 
 | | The FARFAR-MD-NMR ensemble of an HIV-1 TAR excited state | | Descriptor: | The excited state of HIV-1 transactivation response element (31-MER) | | Authors: | Geng, A, Ganser, L, Roy, R, Shi, H, Pratihar, S, Case, D.A, Al-Hashimi, H.M. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA excited conformational state at atomic resolution.
Nat Commun, 14, 2023
|
|
4WNL
 
 | | The X-ray structure of a RNA-binding protein complex | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Singh, N, Blobel, G, Shi, H. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hooking She3p onto She2p for myosin-mediated cytoplasmic mRNA transport.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6LQH
 
 | | High resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6LQJ
 
 | | Low resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
9C2C
 
 | | Cryo-EM structure of the human P2X1 receptor in the NF449-bound inhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4',4'',4'''-{carbonylbis[azanediylbenzene-5,1,3-triylbis(carbonylazanediyl)]}tetra(benzene-1,3-disulfonic acid), P2X purinoceptor 1 | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
9C2A
 
 | | Cryo-EM structure of the human P2X1 receptor in the apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
6WPG
 
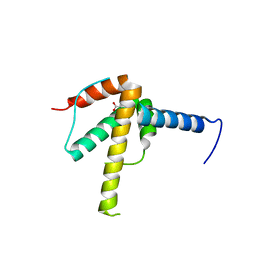 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | Descriptor: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | Authors: | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
9C2B
 
 | | Cryo-EM structure of the human P2X1 receptor in the ATP-bound desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
7BRM
 
 | | Architecture of curli complex | | Descriptor: | Curli production assembly/transport protein CsgG, csgf | | Authors: | Zhang, M, Shi, H. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6VCD
 
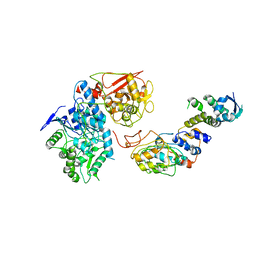 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | Descriptor: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | Authors: | Wang, H, Shi, H, Zheng, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
6UGM
 
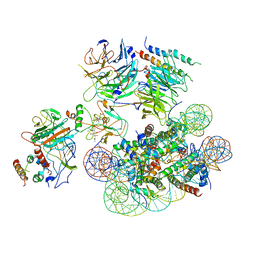 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6UZ3
 
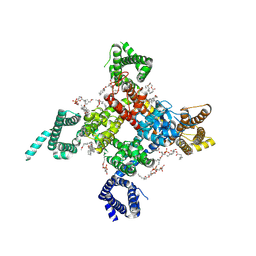 | | Cardiac sodium channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
6UZ0
 
 | | Cardiac sodium channel with flecainide | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
3MD3
 
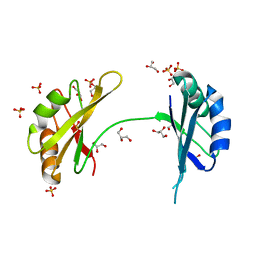 | | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1, SULFATE ION | | Authors: | Li, H, Shi, H, Zhu, Z, Wang, H, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1)
To be published
|
|
3MD1
 
 | | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Li, H, Shi, H, Li, Y, Cui, Y, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1)
To be published
|
|
7V8I
 
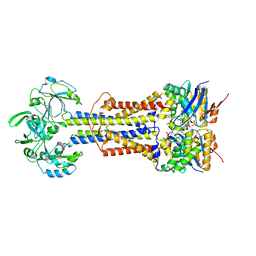 | | LolCD(E171Q)E with bound AMPPNP in nanodiscs | | Descriptor: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7V8M
 
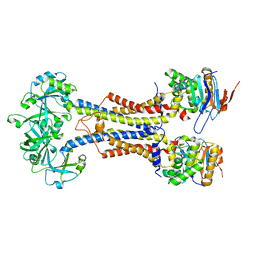 | | LolCDE-apo in nanodiscs | | Descriptor: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Luo, Q.S, Bei, W.W, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7V8L
 
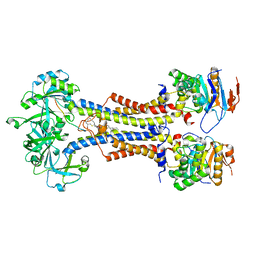 | | LolCDE with bound RcsF in nanodiscs | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | Authors: | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
