5T0U
 
 | | CTCF ZnF2-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T00
 
 | | Human CTCF ZnF3-7 and methylated DNA complex | | Descriptor: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
1WTE
 
 | | Crystal structure of type II restrcition endonuclease, EcoO109I complexed with cognate DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*GP*CP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*GP*CP*CP*CP*GP*GP*T)-3', EcoO109IR, ... | | Authors: | Hashimoto, H, Shimizu, T, Imasaki, T, Kato, M, Shichijo, N, Kita, K, Sato, M. | | Deposit date: | 2004-11-22 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA
J.Biol.Chem., 280, 2005
|
|
1WTD
 
 | | Crystal structure of type II restrcition endonuclease, EcoO109I DNA-free form | | Descriptor: | EcoO109IR, GLYCEROL | | Authors: | Hashimoto, H, Shimizu, T, Imasaki, T, Kato, M, Shichijo, N, Kita, K, Sato, M. | | Deposit date: | 2004-11-22 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA
J.Biol.Chem., 280, 2005
|
|
5XG3
 
 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
3WDR
 
 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide | | Descriptor: | BICARBONATE ION, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
1UZ5
 
 | |
5YD8
 
 | |
2ZHR
 
 | |
1R35
 
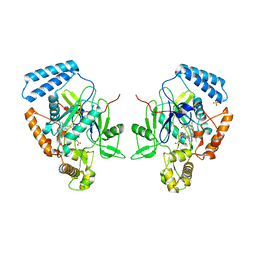 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER, TETRAHYDROBIOPTERIN AND 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE | | Descriptor: | 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Shieh, H.S, Stevens, A.M, Stallings, W.C. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4-Fluorinated L-lysine analogs as selective i-NOS inhibitors: methodology for introducing fluorine into the lysine side chain.
Org.Biomol.Chem., 1, 2003
|
|
2ZO1
 
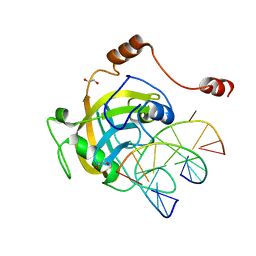 | | Mouse NP95 SRA domain DNA specific complex 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), ... | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
1RFP
 
 | |
1T5F
 
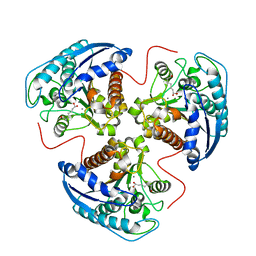 | | arginase I-AOH complex | | Descriptor: | (S)-2-AMINO-7,7-DIHYDROXYHEPTANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Shin, H, Cama, E, Christianson, D.W. | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of amino acid aldehydes as transition-state analogue inhibitors of arginase
J.Am.Chem.Soc., 126, 2004
|
|
3WDQ
 
 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
5XAO
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala in complexes with sodium and chloride ions | | Descriptor: | ACETIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kamitori, S, Sode, K. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
1WUZ
 
 | | Structure of EC1 domain of CNR | | Descriptor: | Pcdha4 protein | | Authors: | Morishita, H, Umitsu, M, Yamaguchi, T, Murata, Y, Shibata, N, Udaka, K, Higuchi, Y, Akutsu, H, Yagi, T, Ikegami, T. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural diversity of the first cadherin domains revealed by the structure of CNR/Protocadherin alpha
To be Published
|
|
2ZHV
 
 | | Crystal structure of BACE1 at pH 7.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHU
 
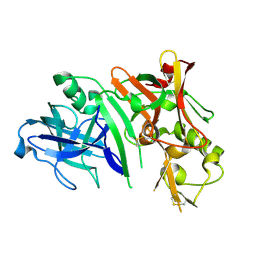 | | Crystal structure of BACE1 at pH 5.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHS
 
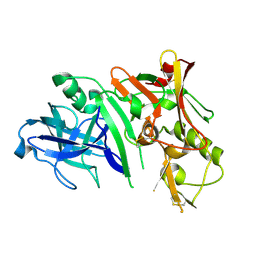 | | Crystal structure of BACE1 at pH 4.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZO0
 
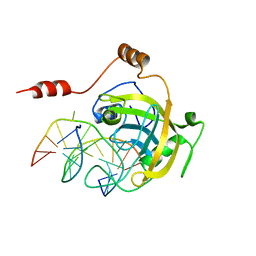 | | mouse NP95 SRA domain DNA specific complex 1 | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZHT
 
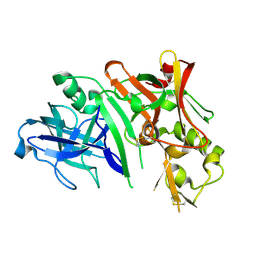 | | Crystal structure of BACE1 at pH 4.5 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
1UIF
 
 | |
1UID
 
 | |
2E6K
 
 | | X-ray structure of Thermus thermopilus HB8 TT0505 | | Descriptor: | Transketolase | | Authors: | Yoshida, H, Kamitori, S, Agari, Y, Iino, H, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structure of Thermus thermophilus HB8 TT0505
To be Published
|
|
3VKM
 
 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
