3VLB
 
 | | Crystal structure of xeg-edgp | | 分子名称: | EDGP, Xyloglucan-specific endo-beta-1,4-glucanase A | | 著者 | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | 登録日 | 2011-11-30 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | 著者 | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.201 Å) | | 主引用文献 | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LS8
 
 | | The monomeric structure of G80A/H81A/H82A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-17 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
2ADV
 
 | | Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | 分子名称: | Glutaryl 7- Aminocephalosporanic Acid Acylase | | 著者 | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | 登録日 | 2005-07-21 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.244 Å) | | 主引用文献 | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AE4
 
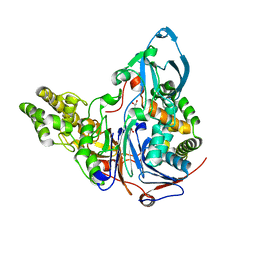 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | 分子名称: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase, SULFATE ION | | 著者 | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | 登録日 | 2005-07-21 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AE3
 
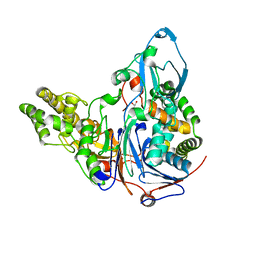 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | 分子名称: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase | | 著者 | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | 登録日 | 2005-07-21 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7YBD
 
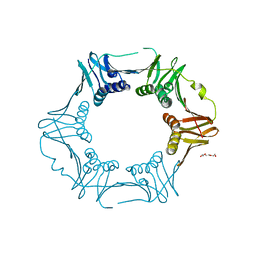 | | Crystal structure of sliding DNA clamp of Clostridioides difficile | | 分子名称: | Beta sliding clamp, TRIETHYLENE GLYCOL | | 著者 | Hishiki, A, Okazaki, S, Hara, K, Hashimoto, H. | | 登録日 | 2022-06-29 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of the sliding DNA clamp from the Gram-positive anaerobic bacterium Clostridioides difficile.
J.Biochem., 173, 2022
|
|
6L2D
 
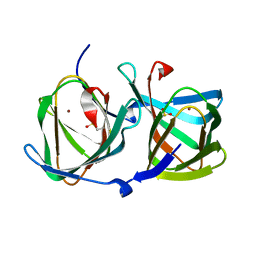 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | 分子名称: | COPPER (II) ION, Cupin_2 domain-containing protein | | 著者 | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.198 Å) | | 主引用文献 | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LTM
 
 | | The dimeric structure of G80A/H81A/H82A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTL
 
 | | The dimeric structure of G80A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6S9J
 
 | | Crystal structure of TfR1 mimicry in complex with GP1 from MACV | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex, ... | | 著者 | Diskin, R, Cohen-Dvashi, H. | | 登録日 | 2019-07-15 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.698 Å) | | 主引用文献 | Rational design of universal immunotherapy for TfR1-tropic arenaviruses.
Nat Commun, 11, 2020
|
|
6UEQ
 
 | |
8YHR
 
 | | DHODH in complex with furocoumavirin | | 分子名称: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | 登録日 | 2024-02-28 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | 著者 | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | 登録日 | 2017-03-14 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.615 Å) | | 主引用文献 | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
6GH8
 
 | | Crystal structure of GP1 domain of Lujo virus in complex with the first CUB domain of neuropilin-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glycoprotein, ... | | 著者 | Cohen-Dvashi, H, Kilimnik, I, Diskin, R. | | 登録日 | 2018-05-06 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural basis for receptor recognition by Lujo virus.
Nat Microbiol, 3, 2018
|
|
8J9Q
 
 | |
8J9R
 
 | |
9ARQ
 
 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-02-23 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARS
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-02-23 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ART
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-02-23 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-03-04 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
5WQW
 
 | | X-ray structure of catalytic domain of autolysin from Clostridium perfringens | | 分子名称: | 1,2-ETHANEDIOL, N-acetylglucosaminidase | | 著者 | Tamai, E, Sekiya, H, Goda, E, Makihata, N, Maki, J, Yoshida, H, Kamitori, S. | | 登録日 | 2016-11-29 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
FEBS Lett., 591, 2017
|
|
2AHL
 
 | | Crystal structure of the hydroxylamine-induced deoxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein | | 分子名称: | CADDIE PROTEIN ORF378, COPPER (I) ION, NITRATE ION, ... | | 著者 | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | 登録日 | 2005-07-28 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
2AHK
 
 | | Crystal structure of the met-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein obtained by soking in cupric sulfate for 6 months | | 分子名称: | CADDIE PROTEIN ORF378, COPPER (II) ION, NITRATE ION, ... | | 著者 | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | 登録日 | 2005-07-28 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
6S8J
 
 | | Structure of ZEBOV GP in complex with 5T0180 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope Glycoprotein 1, Envelope glycoprotein, ... | | 著者 | Diskin, R, Cohen-Dvashi, H. | | 登録日 | 2019-07-10 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
