3HDI
 
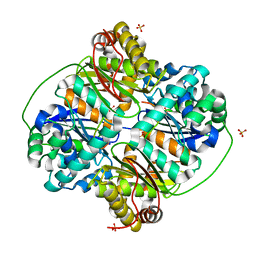 | | Crystal structure of Bacillus halodurans metallo peptidase | | Descriptor: | COBALT (II) ION, Processing protease, SULFATE ION, ... | | Authors: | Aleshin, A, Gramatikova, S, Strongin, A.Y, Stec, B, Liddington, R.C, Smith, J.W. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal and solution structures of a prokaryotic M16B peptidase: an open and shut case.
Structure, 17, 2009
|
|
3ED1
 
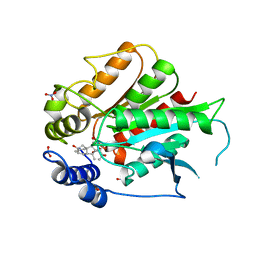 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
3EBL
 
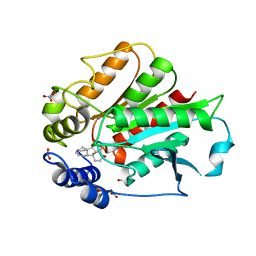 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
4WGY
 
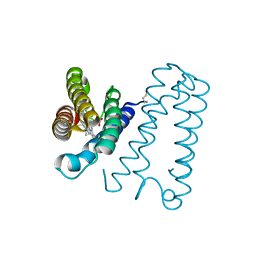 | |
3IZ1
 
 | | C-alpha model fitted into the EM structure of Cx26M34A | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IZ2
 
 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
4S0P
 
 | |
4S0O
 
 | |
3ERD
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH DIETHYLSTILBESTROL AND A GLUCOCORTICOID RECEPTOR INTERACTING PROTEIN 1 NR BOX II PEPTIDE | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIETHYLSTILBESTROL, ... | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
3JSV
 
 | | Crystal structure of mouse NEMO CoZi in complex with Lys63-linked di-ubiquitin | | Descriptor: | NF-kappa-B essential modulator, Ubiquitin | | Authors: | Yoshikawa, A, Sato, Y, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-09-11 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin
Febs Lett., 583, 2009
|
|
5DBH
 
 | |
3ERT
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH 4-HYDROXYTAMOXIFEN | | Descriptor: | 4-HYDROXYTAMOXIFEN, PROTEIN (ESTROGEN RECEPTOR ALPHA) | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-30 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
6KCS
 
 | | Crystal structure of HIRAN domain of HLTF in complex with duplex DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*AP*CP*GP*TP*AP*CP*AP*GP*T)-3'), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of HIRAN domain of human HLTF bound to duplex DNA provides structural basis for DNA unwinding to initiate replication fork regression.
J.Biochem., 167, 2020
|
|
8IYP
 
 | | Crystal structure of serine palmitoyltransferase soaked in 190 mM D-serine solution | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
8IYT
 
 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
3FLM
 
 | | Crystal structure of menD from E.coli | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Priyadarshi, A, Hwang, K.Y. | | Deposit date: | 2008-12-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
8WAF
 
 | | Crystal structure of the C-terminal fragment (residues 756-982 with the C864S mutation) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Takano, A, Nakamura, Y, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
8WAG
 
 | | Crystal structure of the C-terminal fragment (residues 716-982) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Nakamura, Y, Takano, A, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
3W0W
 
 | | The complex between T36-5 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide in space group P212121 | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXS
 
 | | The complex between H27-14 TCR and HLA-A24 bound to HIV-1 Nef134-10(6L) peptide | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, H27-14 TCR alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXP
 
 | | HLA-A24 in complex with HIV-1 Nef134-10(6L) | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXT
 
 | | T36-5 TCR specific for HLA-A24-Nef134-10 | | Descriptor: | T36-5 TCR alpha chain, T36-5 TCR beta chain | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXR
 
 | | The complex between H27-14 TCR and HLA-A24 bound to HIV-1 Nef134-10(wt) peptide | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, H27-14 TCR alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
4H4T
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 T176R mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
4H4V
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/T176R/Q177G mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
