3QCD
 
 | |
3QCI
 
 | |
3QCJ
 
 | |
3QCB
 
 | | Human receptor protein tyrosine phosphatase gamma, domain 1, apo | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecule receptor protein tyrosine phosphatase [gamma](RPTP[gamma]) ligands that inhibit phosphatase activity via perturbation of the tryptophan-proline-aspartate (WPD) loop
J.Med.Chem., 54, 2011
|
|
3QCN
 
 | | Human receptor protein tyrosine phosphatase gamma, domain 1, trigonal crystal form | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Small molecule receptor protein tyrosine phosphatase [gamma](RPTP[gamma]) ligands that inhibit phosphatase activity via perturbation of the tryptophan-proline-aspartate (WPD) loop
J.Med.Chem., 54, 2011
|
|
5TWN
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
5TWM
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
5TRH
 
 | |
5TRI
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid, NS5B RNA-DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and initial optimization of alkoxyanthranilic acid derivatives as inhibitors of HCV NS5B polymerase.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TRJ
 
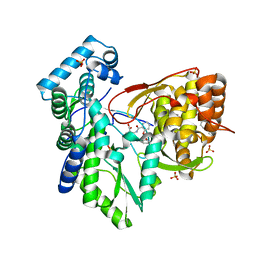 | |
5TRK
 
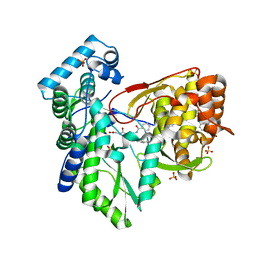 | |
5DMJ
 
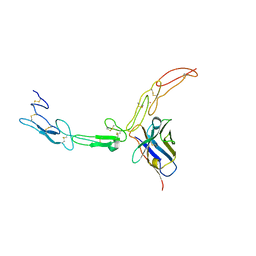 | |
5DMI
 
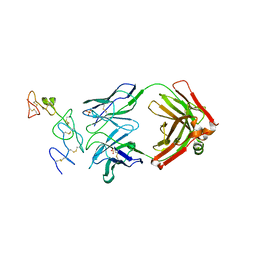 | |
3Q0Z
 
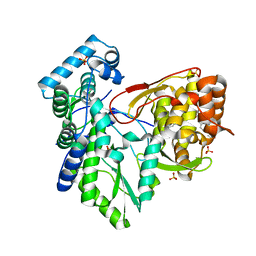 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Syntheses and initial evaluation of a series of indolo-fused heterocyclic inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGF
 
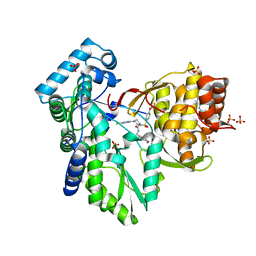 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGG
 
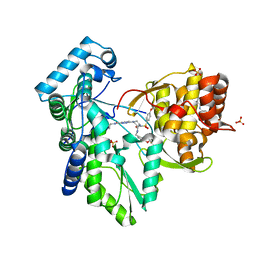 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGI
 
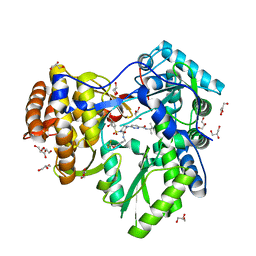 | |
3QGH
 
 | |
3QGE
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5CIR
 
 | | Crystal structure of death receptor 4 (DR4; TNFFRSF10A) bound to TRAIL (TNFSF10) | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 10, Tumor necrosis factor receptor superfamily member 10A, ... | | Authors: | Sheriff, S. | | Deposit date: | 2015-07-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the death receptor 4-TNF-related apoptosis-inducing ligand (DR4-TRAIL) complex.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
5E8Y
 
 | |
5E8W
 
 | |
5E8U
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F,S280T, Y282F,S287N,A350C,L352F) | | Descriptor: | GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E91
 
 | |
