3DJ3
 
 | |
3DIW
 
 | | c-terminal beta-catenin bound TIP-1 structure | | Descriptor: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
1T17
 
 | | Solution Structure of the 18 kDa Protein CC1736 from Caulobacter crescentus: The Northeast Structural Genomics Consortium Target CcR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Shen, Y, Atreya, H.S, Acton, T, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity.
Proteins, 58, 2005
|
|
5C54
 
 | |
5C55
 
 | |
1XFX
 
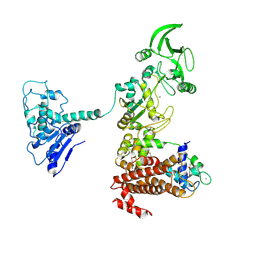 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 10 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFZ
 
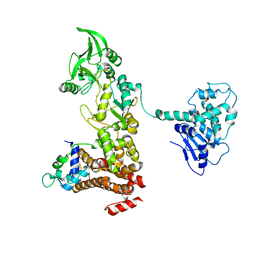 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 1 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
3COJ
 
 | |
1XFU
 
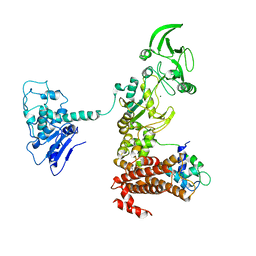 | | Crystal structure of anthrax edema factor (EF) truncation mutant, EF-delta 64 in complex with calmodulin | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XN7
 
 | | Solution Structure of E.Coli Protein yhgG: The Northeast Structural Genomics Consortium Target ET95 | | Descriptor: | Hypothetical protein yhgG | | Authors: | Shen, Y, Acton, T, Atreya, H.S, Ma, L, Liu, G, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of E.Coli Protein yhgG: The Northeast Structural Genomics Consortium Target ET95
To be Published
|
|
1XFY
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFW
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3'5' cyclic AMP (cAMP) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1Y0V
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and pyrophosphate | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.-J. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor
Embo J., 24, 2005
|
|
1W93
 
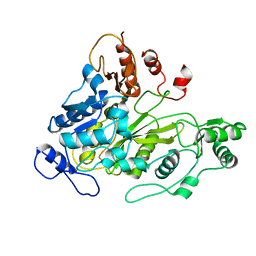 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-Coenzyme A Carboxylase from Saccharomyces cerevisiae | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1W96
 
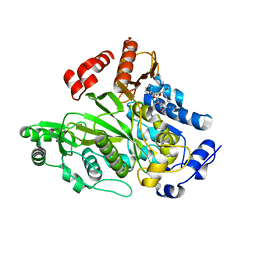 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-coenzyme A Carboxylase from Saccharomyces cerevisiae in Complex with Soraphen A | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, SORAPHEN A | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
3SLU
 
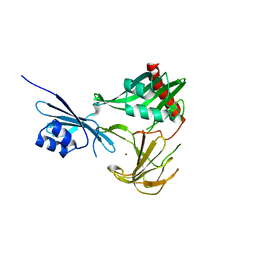 | | Crystal structure of NMB0315 | | Descriptor: | M23 peptidase domain protein, NICKEL (II) ION | | Authors: | Shen, Y, Wang, X, Yang, X, Xu, H. | | Deposit date: | 2011-06-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
5WDE
 
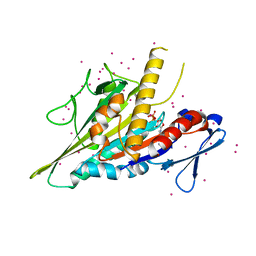 | | Crystal structure of the KIFC3 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC3, MAGNESIUM ION, ... | | Authors: | Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
3SUZ
 
 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
2G48
 
 | |
2G47
 
 | |
3UB3
 
 | | D96N variant of TIR domain of Mal/TIRAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Shen, Y, Lin, Z. | | Deposit date: | 2011-10-23 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
2G49
 
 | |
3SV1
 
 | | Crystal structure of APP peptide bound rat Mint2 PARM | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2, Amyloid beta A4 protein | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
2G54
 
 | |
3UB2
 
 | | TIR domain of Mal/TIRAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Shen, Y, Lin, Z. | | Deposit date: | 2011-10-23 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
