3LXX
 
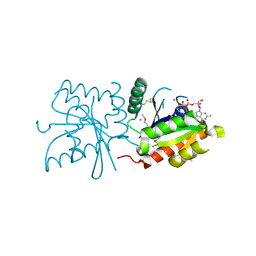 | | Crystal structure of human GTPase IMAP family member 4 | | Descriptor: | GTPase IMAP family member 4, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shen, Y, Nedyalkova, L, Tong, Y, Tempel, W, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human GTPase IMAP family member 4
to be published
|
|
3DJ3
 
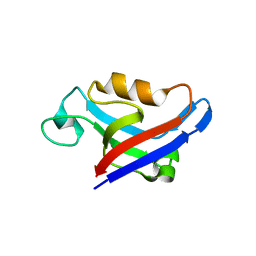 | |
3DIW
 
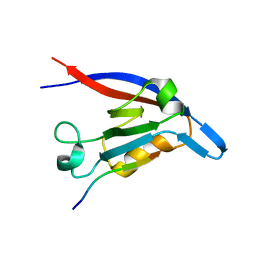 | | c-terminal beta-catenin bound TIP-1 structure | | Descriptor: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
1W96
 
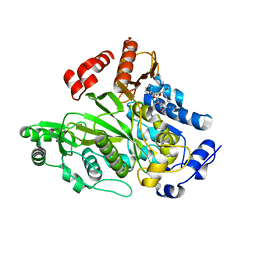 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-coenzyme A Carboxylase from Saccharomyces cerevisiae in Complex with Soraphen A | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, SORAPHEN A | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
2R7I
 
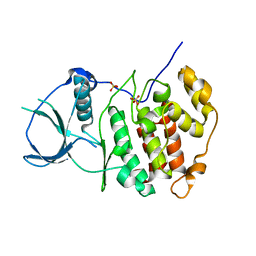 | | Crystal structure of catalytic subunit of protein kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
2R6M
 
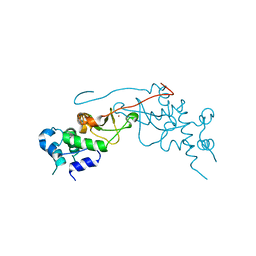 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
2G47
 
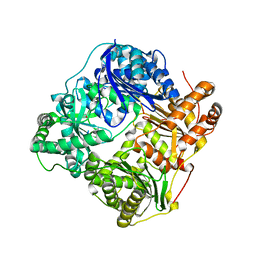 | |
2G48
 
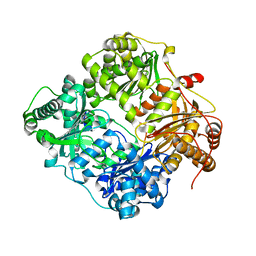 | |
2G49
 
 | |
2G56
 
 | |
3EAP
 
 | | Crystal structure of the RhoGAP domain of ARHGAP11A | | Descriptor: | Rho GTPase-activating protein 11A, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the RhoGAP domain of ARHGAP11A
To be Published
|
|
1PK0
 
 | | Crystal Structure of the EF3-CaM complexed with PMEApp | | Descriptor: | (ADENIN-9-YL-ETHOXYMETHYL)-HYDROXYPHOSPHINYL-DIPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Shen, Y, Tang, W.J. | | Deposit date: | 2003-06-04 | | Release date: | 2004-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1NR3
 
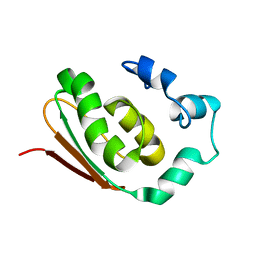 | | SOLUTION STRUCTURE OF THE PROTEIN MTH0916: THE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT212 | | Descriptor: | DNA-binding protein tfx | | Authors: | Shen, Y, Liu, G, Bhaskaran, R, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-23 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE PROTEIN MTH0916: THE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT212
To be Published
|
|
3JZY
 
 | | Crystal structure of human Intersectin 2 C2 domain | | Descriptor: | Intersectin 2, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tempel, W, Tong, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of human Intersectin 2 C2 domain
To be Published
|
|
3GF9
 
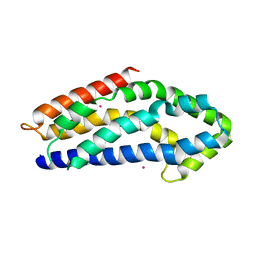 | | Crystal structure of human Intersectin 2 RhoGEF domain | | Descriptor: | Intersectin 2, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Tempel, W, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Intersectin 2 RhoGEF domain
To be Published
|
|
1S26
 
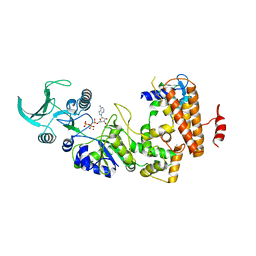 | | Structure of Anthrax Edema Factor-Calmodulin-alpha,beta-methyleneadenosine 5'-triphosphate Complex Reveals an Alternative Mode of ATP Binding to the Catalytic Site | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Bohm, A, Tang, W.-J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of anthrax edema factor-calmodulin-adenosine-5'-(alpha,beta-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site
Biochem.Biophys.Res.Commun., 317, 2004
|
|
3KH0
 
 | | Crystal structure of the Ras-association (RA) domain of RALGDS | | Descriptor: | Ral guanine nucleotide dissociation stimulator, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tempel, W, Wang, H, Tong, Y, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Ras-association (RA) domain of RALGDS
to be published
|
|
3DZ8
 
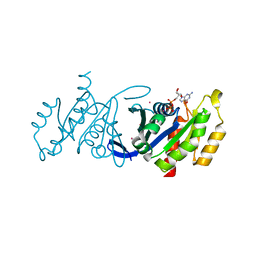 | | Crystal structure of human Rab3B GTPase bound with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-3B, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Sukumar, D, Tempel, W, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Wilkstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Rab3B GTPase bound with GDP
To be Published
|
|
3KKS
 
 | |
3GJ9
 
 | | crystal structure of TIP-1 in complex with c-terminal of Kir2.3 | | Descriptor: | C-terminal peptide from Inward rectifier potassium channel 4, CHLORIDE ION, Tax1-binding protein 3, ... | | Authors: | Shen, Y. | | Deposit date: | 2009-03-08 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of inward rectifier potassium channel 2.3 regulation by tax-interacting protein-1
J.Mol.Biol., 392, 2009
|
|
3DJ1
 
 | | crystal structure of TIP-1 wild type | | Descriptor: | SULFATE ION, Tax1-binding protein 3 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3I6D
 
 | | Crystal structure of PPO from bacillus subtilis with AF | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Shen, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis
J.Struct.Biol., 170, 2010
|
|
1LVC
 
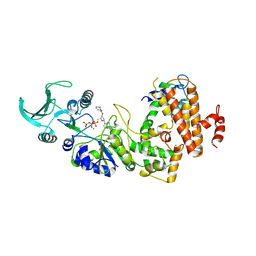 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 2' deoxy, 3' anthraniloyl ATP | | Descriptor: | 3'ANTHRANILOYL-2'-DEOXY-ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, YTTERBIUM (III) ION, ... | | Authors: | Shen, Y, Lee, Y.-S, Soelaiman, S, Bergson, P, Lu, D, Chen, A, Beckingham, K, Grabarek, Z, Mrksich, M, Tang, W.-J. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins
Embo J., 21, 2002
|
|
3KKR
 
 | |
1CRW
 
 | |
