2F7S
 
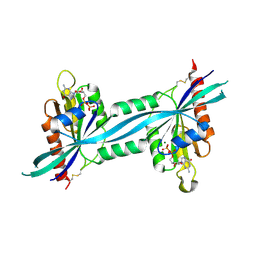 | | The crystal structure of human Rab27b bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Wang, J, Ismail, S, Shen, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of human Rab27b
To be Published
|
|
2YD4
 
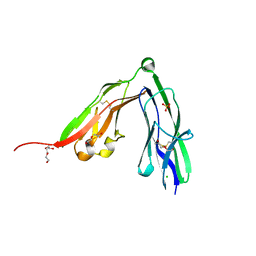 | | Crystal structure of the N-terminal Ig1-2 module of Chicken Receptor Protein Tyrosine Phosphatase Sigma | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PROTEIN-TYROSINE PHOSPHATASE CRYPALPHA1 ISOFORM, ... | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD1
 
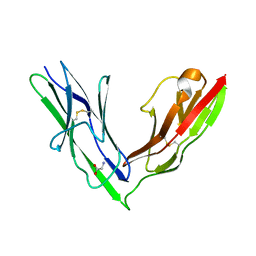 | | Crystal structure of the N-terminal Ig1-2 module of Drosophila Receptor Protein Tyrosine Phosphatase DLAR | | Descriptor: | GLYCINE, TYROSINE-PROTEIN PHOSPHATASE LAR | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD7
 
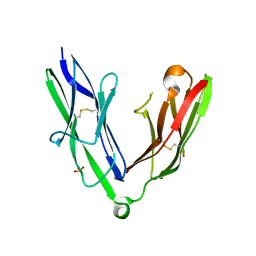 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Delta | | Descriptor: | PHOSPHATE ION, PTPRD PROTEIN | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD5
 
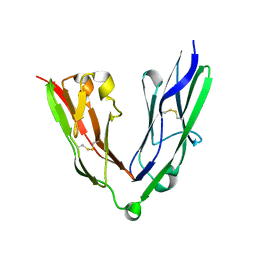 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR | | Descriptor: | RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE F | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD3
 
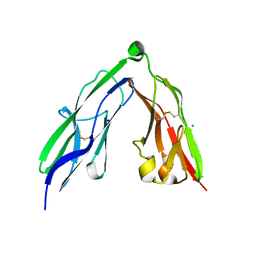 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Sigma | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE S, SODIUM ION | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD2
 
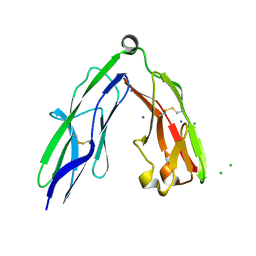 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Sigma | | Descriptor: | CHLORIDE ION, IODIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE S | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
6VX5
 
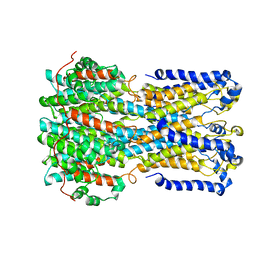 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
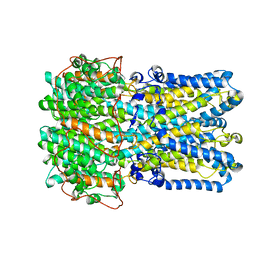 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
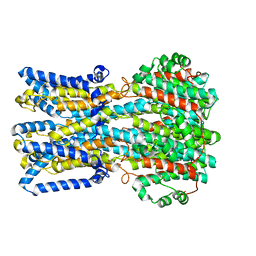 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
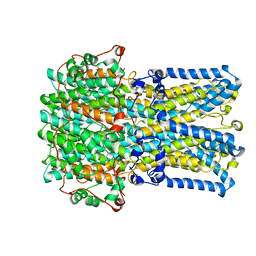 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX7
 
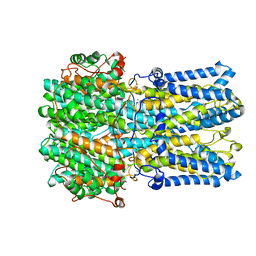 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2YD8
 
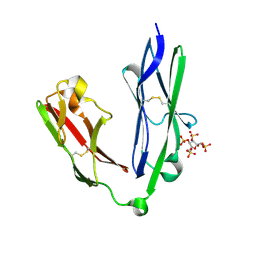 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR in complex with sucrose octasulphate | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE F | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-18 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
4ZRZ
 
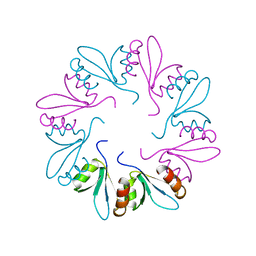 | | PlyCB mutant R66E | | Descriptor: | PlyCB | | Authors: | Gallagher, D.T, Nelson, D.C, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A bacteriophage endolysin that eliminates intracellular streptococci.
Elife, 5, 2016
|
|
4ZRI
 
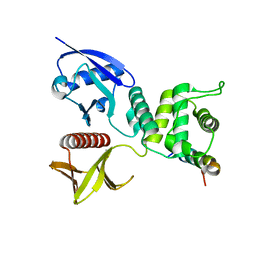 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
3GXA
 
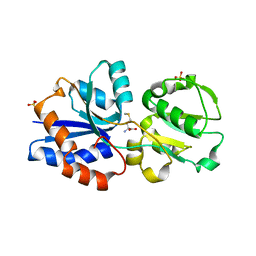 | | Crystal structure of GNA1946 | | Descriptor: | METHIONINE, Outer membrane lipoprotein GNA1946, SULFATE ION | | Authors: | Yang, X, Shen, Y. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of lipoprotein GNA1946 from Neisseria meningitidis
J.Struct.Biol., 168, 2009
|
|
4ZRJ
 
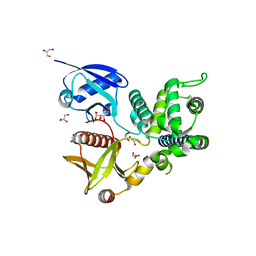 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
3IR1
 
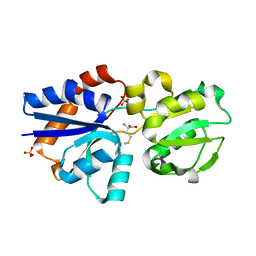 | | Crystal Structure of Lipoprotein GNA1946 from Neisseria meningitidis | | Descriptor: | METHIONINE, Outer membrane lipoprotein GNA1946, SULFATE ION | | Authors: | Yang, X, Wu, Z, Wang, X, Shen, Y. | | Deposit date: | 2009-08-21 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of lipoprotein GNA1946 from Neisseria meningitidis
J.Struct.Biol., 168, 2009
|
|
3O59
 
 | |
3Q5M
 
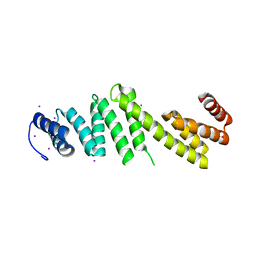 | | Crystal structure of Escherichia coli BamD | | Descriptor: | IODIDE ION, UPF0169 lipoprotein yfiO | | Authors: | Dong, C, Hou, H, Yang, X, Dong, Y, Shen, Y. | | Deposit date: | 2010-12-28 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4O9B
 
 | | The Structure of CC1-IH in human STIM1. | | Descriptor: | CADMIUM ION, Stromal interaction molecule 1 | | Authors: | Cui, B, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1.
Plos One, 8, 2013
|
|
5EWT
 
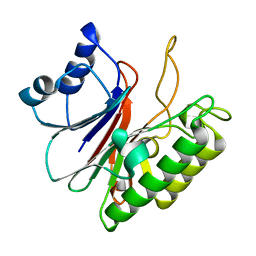 | |
6W6D
 
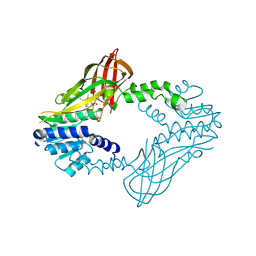 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
4NSD
 
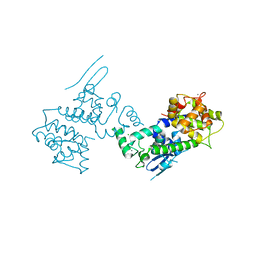 | | Crystal Structure of CBARA1 in the Ca2+ Binding Form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, L, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake.
Embo J., 33, 2014
|
|
8W6G
 
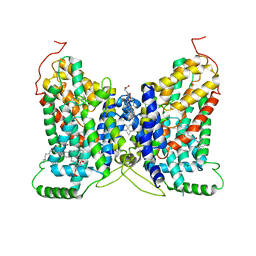 | | NaDC1 with inhibitor ACA | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-[[(~{E})-3-(4-pentylphenyl)prop-2-enoyl]amino]benzoic acid, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
