6IHC
 
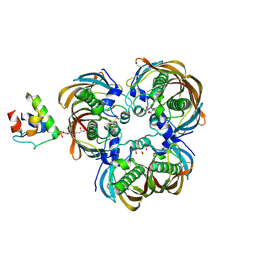 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) Y100A mutant in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, CITRIC ACID, N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, ... | | Authors: | Shen, S.Q, Zhang, L, Zhang, L. | | Deposit date: | 2018-09-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A back-door Phenylalanine coordinates the stepwise hexameric loading of acyl carrier protein by the fatty acid biosynthesis enzyme beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
Int. J. Biol. Macromol., 128, 2019
|
|
8JZW
 
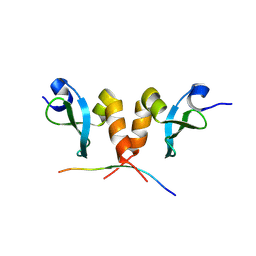 | |
8IKH
 
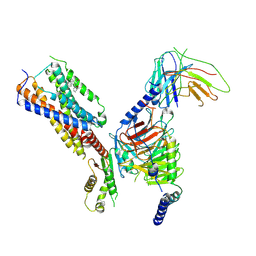 | | Cryo-EM structure of human receptor with G proteins | | Descriptor: | 3-[(1R)-1-(2-methoxyphenyl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Shao, Z.H. | | Deposit date: | 2023-02-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8IKG
 
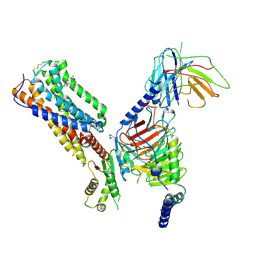 | | Cryo-EM structure of human receptor with G proteins | | Descriptor: | 3-[(1S)-1-(furan-2-yl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Shao, Z.H. | | Deposit date: | 2023-02-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZJE
 
 | | Cryo-EM structure of kisspeptin receptor bound to TAK-448 | | Descriptor: | ACY-DTY-HYP-ASN-THR-PHE-XZA-LEU-NMM-TRP-NH2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, S, Liu, H, Xu, H.E. | | Deposit date: | 2024-05-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of kisspeptin receptor bound to TAK-448
To Be Published
|
|
8ZJD
 
 | | Cryo-EM structure of kisspeptin receptor bound to KP-10 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Shen, S, Liu, H, Xu, H.E. | | Deposit date: | 2024-05-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of kisspeptin receptor bound to KP-10
To Be Published
|
|
7EIU
 
 | | Crystal structure of Mei2 RRM3 in complex with 8mer meiRNA | | Descriptor: | GLYCEROL, Meiosis protein mei2, RNA (5'-R(P*UP*UP*CP*UP*GP*C)-3') | | Authors: | Shen, S.Y, Lv, M.Q. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
8K6N
 
 | | Structural complex of neuropeptide Y receptor 2 | | Descriptor: | C-flanking peptide of NPY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, S.Y, Zhao, C, Shao, Z.H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural complex of neuropeptide Y receptor 2
To Be Published
|
|
8K6M
 
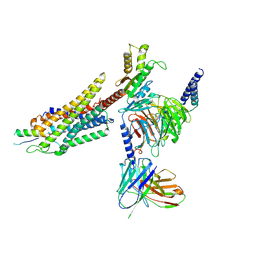 | | Structural complex of neuropeptide Y receptor 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Shen, S.Y, Zhao, C, Shao, Z.H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural complex of neuropeptide Y receptor 1
To Be Published
|
|
8K6O
 
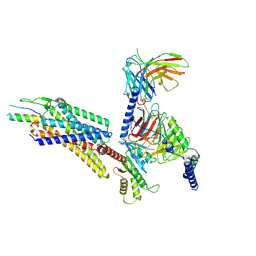 | | Structural complex of neuropeptide Y receptor 1 | | Descriptor: | C-flanking peptide of NPY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, S.Y, Zhao, C, Shao, Z.H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural complex of neuropeptide Y receptor 1
To Be Published
|
|
7EIO
 
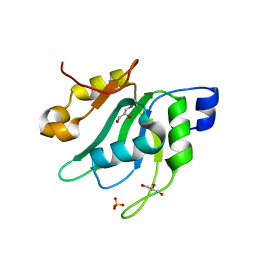 | | Crystal Structure of Mei2 RRM3 | | Descriptor: | GLYCEROL, Meiosis protein mei2, SULFATE ION | | Authors: | Shen, S.Y, Li, F.D. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
8GS9
 
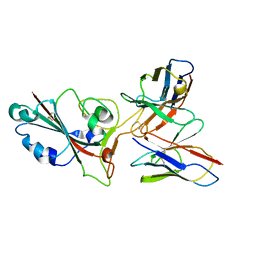 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | Descriptor: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | Authors: | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
8GSB
 
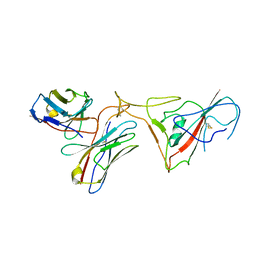 | |
7LFM
 
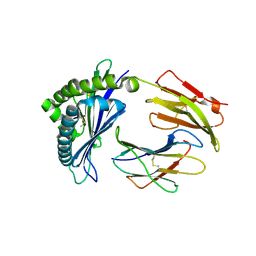 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFL
 
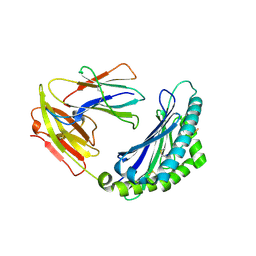 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, MONOCLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFI
 
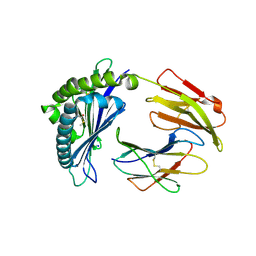 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFJ
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, ALA MUTANT, REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
7LFK
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, THR MUTANT, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
6J23
 
 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
8EZ1
 
 | | Human Ornithine Aminotransferase (hOAT) co-crystallized with its inactivator 3-Amino-4-fluorocyclopentenecarboxylic Acid | | Descriptor: | (1R,3S,4Z)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, (3E,4E)-4-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-iminocyclopent-1-ene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Shen, S, Silverman, R, Liu, D. | | Deposit date: | 2022-10-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Mechanistic Basis for the Inactivation of Human Ornithine Aminotransferase by (3 S ,4 S )-3-Amino-4-fluorocyclopentenecarboxylic Acid.
Molecules, 28, 2023
|
|
6AJK
 
 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
7LK0
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | Descriptor: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LK1
 
 | | Ornithine Aminotransferase (OAT) with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) - 1 Hour Soaking | | Descriptor: | (1R,4R)-4-fluoro-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-2-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
