3ZIM
 
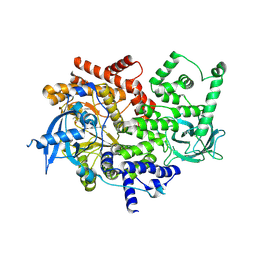 | | Discovery of a potent and isoform-selective targeted covalent inhibitor of the lipid kinase PI3Kalpha | | Descriptor: | 1-[4-[[2-(1H-indazol-4-yl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-6-methyl-hept-5-ene-1,4- dione, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Nacht, M, Qiao, L, Sheets, M.P, Martin, T.S, Labenski, M, Mazdiyasni, H, Karp, R, Zhu, Z, Chaturvedi, P, Bhavsar, D, Niu, D, Westlin, W, Petter, R.C, Medikonda, A.P, Jestel, A, Blaesse, M, Singh, J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent and Isoform-Selective Targeted Covalent Inhibitor of the Lipid Kinase Pi3Kalpha
J.Med.Chem., 56, 2013
|
|
7L5J
 
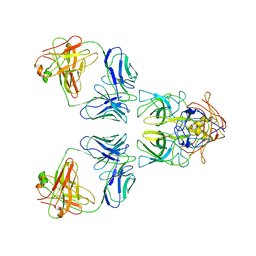 | |
6HRJ
 
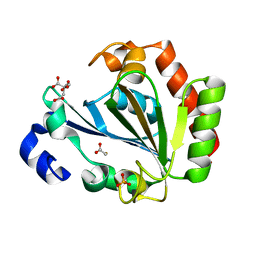 | | Apo - YndL | | Descriptor: | CITRATE ANION, SULFATE ION, YndL, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
5OJQ
 
 | | The modeled structure of of wild type extended type VI secretion system sheath/tube complex in vibrio cholerae based on cryo-EM reconstruction of the non-contractile sheath/tube complex | | Descriptor: | Haemolysin co-regulated protein, Type VI secretion protein, VipA | | Authors: | Wang, J, Brackmann, M, Castano-Diez, D, Kudryashev, M, Goldie, K, Maier, T, Stahlberg, H, Basler, M. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Nat Microbiol, 2, 2017
|
|
5CR6
 
 | | Structure of pneumolysin at 1.98 A resolution | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
5CQG
 
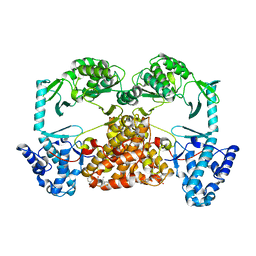 | | Structure of Tribolium telomerase in complex with the highly specific inhibitor BIBR1532 | | Descriptor: | 2-{[(2E)-3-(naphthalen-2-yl)but-2-enoyl]amino}benzoic acid, Telomerase reverse transcriptase | | Authors: | Bryan, C, Rice, C, Hoffman, H, Harkisheimer, M, Sweeney, M, Skordalakes, E. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Telomerase Inhibition by the Highly Specific BIBR1532.
Structure, 23, 2015
|
|
5CR8
 
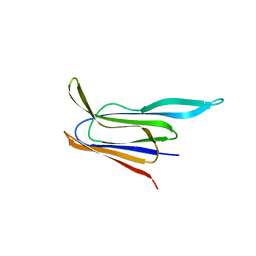 | | Structure of the membrane-binding domain of pneumolysin | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
3SRK
 
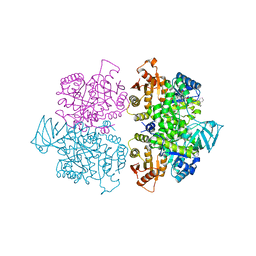 | | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, POTASSIUM ION, Pyruvate kinase, ... | | Authors: | Morgan, H.P, Walsh, M, Blackburn, E.A, Wear, M.A, Boxer, M, Shen, M, McNae, I.W, Michels, P.A.M, Auld, D.S, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase
To be Published
|
|
1ZY6
 
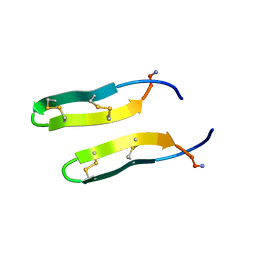 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
3T64
 
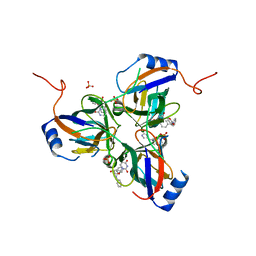 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-[(diphenylmethyl)amino]uridine, 5'-(BENZHYDRYLAMINO)-2',5'-DIDEOXYURIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
3T6Y
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-{[(R)-(1-methyl-1H-imidazol-2-yl)(phenyl)methyl]amino}uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
5OWO
 
 | | Human cytoplasmic Dynein N-Terminus dimerization domain at 1.8 Angstrom resolution | | Descriptor: | CALCIUM ION, Cytoplasmic dynein 1 heavy chain 1, GLYCEROL, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
3T70
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-[(diphenylmethyl)(methyl)amino]uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
3T60
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-(tritylamino)uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
4JBI
 
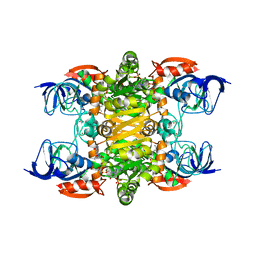 | | 2.35A resolution structure of NADPH bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
4JBG
 
 | | 1.75A resolution structure of a thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
6HRI
 
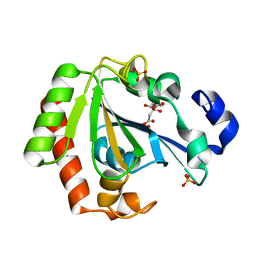 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
3R30
 
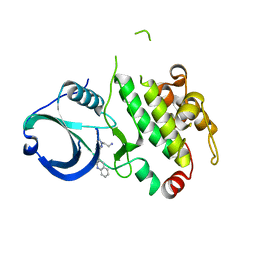 | | MK2 kinase bound to Compound 2 | | Descriptor: | 1-(2-aminoethyl)-3-[2-(quinolin-3-yl)pyridin-4-yl]-1H-pyrazole-5-carboxylic acid, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Fisher, M. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RIN
 
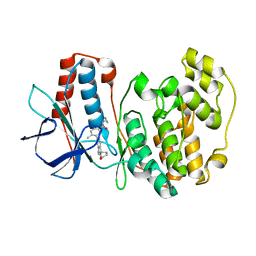 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-(2'-oxo-1',2'-dihydrospiro[cyclopentane-1,3'-indol]-6'-yl)benzamide | | Authors: | Segarra, V, Eastwood, P, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Indolin-2-one p38(alpha) inhibitors I: design, profiling and crystallographic binding mode.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2YM6
 
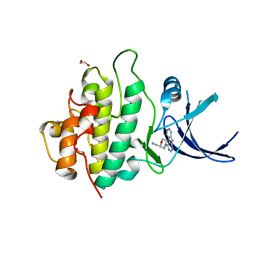 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-4-(9H-pyrido[4',3':4,5]pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]methanamine, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2YM5
 
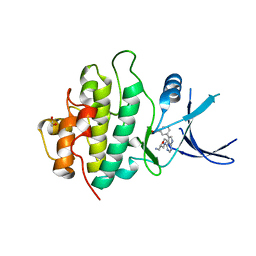 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (3-{4-[(2S)-2-(AMINOMETHYL)MORPHOLIN-4-YL]-7H-PYRROLO[2,3-D]PYRIMIDIN-5-YL}PHENYL)METHANOL, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2YM3
 
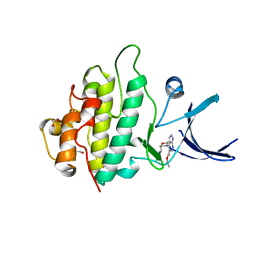 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, ETHYL 4-(2-(AMINOMETHYL)MORPHOLINO)-1H-PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXYLATE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2YM8
 
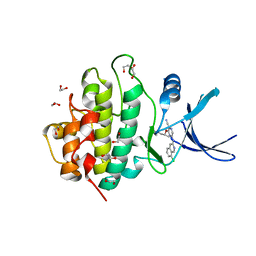 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (R)-5-(8-CHLOROISOQUINOLIN-3-YLAMINO)-3-(1-(DIMETHYLAMINO)PROPAN-2-YLOXY)PYRAZINE-2-CARBONITRILE, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2YM4
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1, ethyl 4-[(2R)-2-(aminomethyl)morpholin-4-yl]-3-(3-cyanophenyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxylate | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
3S3I
 
 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(3-tert-butyl[1,2,4]triazolo[4,3-a]pyridin-7-yl)-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Segarra, V, Aiguade, J, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-05-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel triazolopyridylbenzamides as potent and selective p38 alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
