3UEY
 
 | |
3UGD
 
 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta
Biophys.J., 103, 2012
|
|
3UEJ
 
 | |
3UGL
 
 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | PHOSPHATE ION, Proteine kinase C delta type, ZINC ION, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.357 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
3UFF
 
 | |
3UGI
 
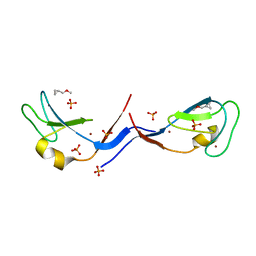 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | (methoxymethyl)cyclopropane, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
7NFX
 
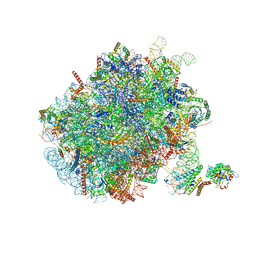 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
6S0K
 
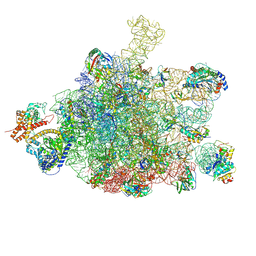 | | Ribosome nascent chain in complex with SecA | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Jomaa, A, Wang, S, Shan, S, Ban, N. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The molecular mechanism of cotranslational membrane protein recognition and targeting by SecA.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7D5Z
 
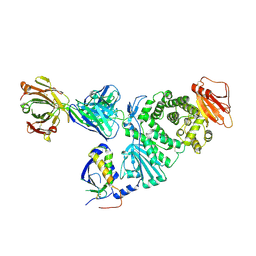 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | Authors: | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
6M0J
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
2XXA
 
 | | The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) | | Descriptor: | 4.5S RNA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ataide, S.F, Schmitz, N, Shen, K, Ke, A, Shan, S, Doudna, J.A, Ban, N. | | Deposit date: | 2010-11-09 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor.
Science, 331, 2011
|
|
2OG2
 
 | | Crystal structure of chloroplast FtsY from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, MALONATE ION, Putative signal recognition particle receptor | | Authors: | Chartron, J, Chandrasekar, S, Ampornpan, P.J, Shan, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chloroplast Signal Recognition Particle (SRP) Receptor: Domain Arrangement Modulates SRP-Receptor Interaction.
J.Mol.Biol., 375, 2007
|
|
7QWQ
 
 | | Ternary complex of ribosome nascent chain with SRP and NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7QWR
 
 | | Structure of the ribosome-nascent chain containing an ER signal sequence in complex with NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7QWS
 
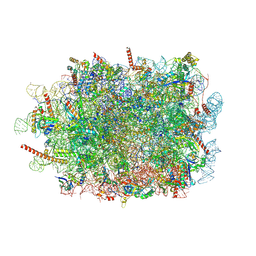 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | Authors: | Zhang, L, Wang, X, Shan, S, Zhang, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | Authors: | Zhang, L, Wang, X, Zhang, S, Shan, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
5AKA
 
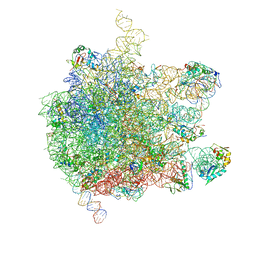 | | EM structure of ribosome-SRP-FtsY complex in closed state | | Descriptor: | 23S ribosomal RNA, 4.5S ribosomal RNA, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | von Loeffelholz, O, Jiang, Q, Ariosa, A, Karuppasamy, M, Huard, K, Berger, I, Shan, S, Schaffitzel, C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Ribosome-Srp-Ftsy Cotranslational Targeting Complex in the Closed State.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6AE8
 
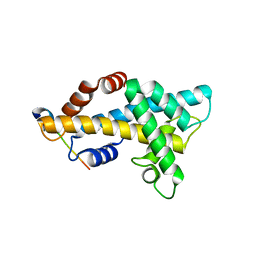 | | Structure insight into histone chaperone Chz1-mediated H2A.Z recognition and replacement | | Descriptor: | BICINE, Histone H2A.Z-specific chaperone CHZ1, Histone H2B.1,Histone H2A.Z | | Authors: | Wang, Y.Y, Shan, S, Zhou, Z. | | Deposit date: | 2018-08-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement.
Plos Biol., 17, 2019
|
|
5Z78
 
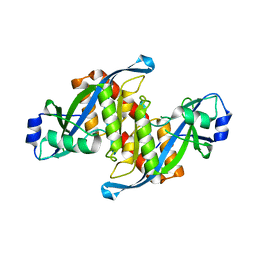 | | Structure of TIRR/53BP1 complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Dai, Y.X, Shan, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR
Nat Commun, 9, 2018
|
|
7WLP
 
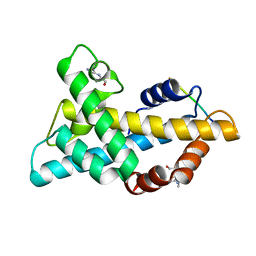 | |
3JYI
 
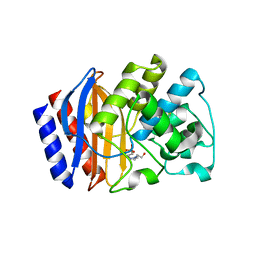 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
5IMQ
 
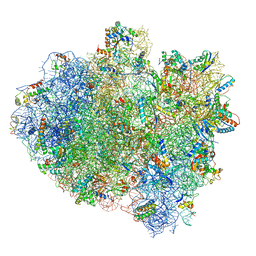 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
8UNH
 
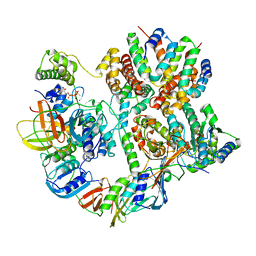 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
