8BP8
 
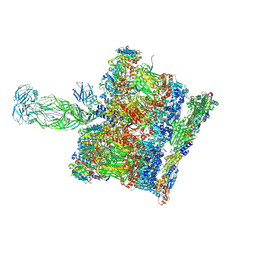 | | SPA of Trypsin untreated Rotavirus TLP spike | | Descriptor: | CALCIUM ION, Inner capsid protein VP2, Intermediate capsid protein VP6, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
8COA
 
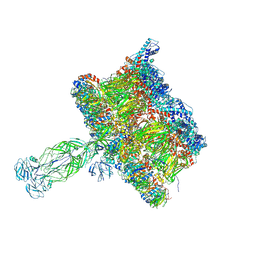 | | in situ Subtomogram average of Immature Rotavirus TLP spike | | Descriptor: | Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, Outer capsid protein VP4 | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
8CO6
 
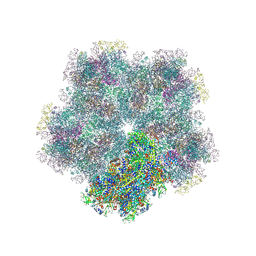 | | Subtomogram average of Immature Rotavirus TLP penton | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
2P6J
 
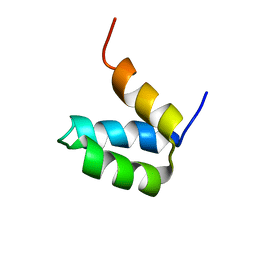 | | Full-sequence computational design and solution structure of a thermostable protein variant | | Descriptor: | designed engrailed homeodomain variant UVF | | Authors: | Shah, P.S, Hom, G.K, Ross, S.A, Lassila, J.K, Crowhurst, K.A, Mayo, S.L. | | Deposit date: | 2007-03-18 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Full-sequence Computational Design and Solution Structure of a Thermostable Protein Variant.
J.Mol.Biol., 372, 2007
|
|
7UH4
 
 | | LXG-associated alpha-helical protein D2 (LapD2) | | Descriptor: | LXG-associated alpha-helical protein D2, SULFATE ION | | Authors: | Klein, T.A, Grebenc, D.W, Shah, P.Y, McArthur, O.D, Surette, M.G, Kim, Y, Whitney, J.C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.
Mbio, 13, 2022
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4XEE
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-23 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4GRV
 
 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
6Z43
 
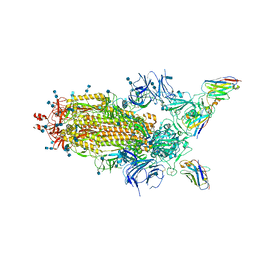 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z97
 
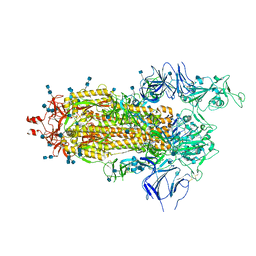 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
8GMH
 
 | | Crystal Structure of the ternary complex of TelA-LXG, LapA3, and LapA4 | | Descriptor: | 1,2-ETHANEDIOL, LXG domain-containing protein, LapA3, ... | | Authors: | Klein, T.A, Shah, P.Y, Gkragkopoulou, P, Grebenc, D.W, Kim, Y, Whitney, J.C. | | Deposit date: | 2023-03-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a tripartite protein complex that targets toxins to the type VII secretion system.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6P9W
 
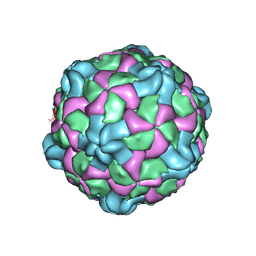 | | Poliovirus (Type 1 Mahoney), receptor catalysed 135S particle map | | Descriptor: | VP1, VP2, VP3 | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
6P9O
 
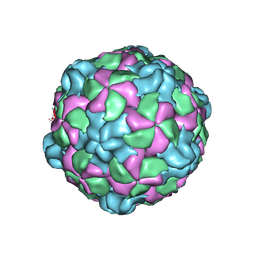 | |
6PSZ
 
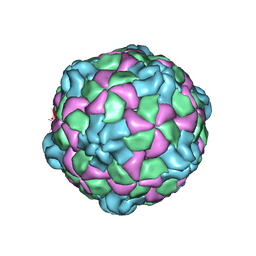 | | Poliovirus (Type 1 Mahoney), heat-catalysed 135S particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-07-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
6Q0B
 
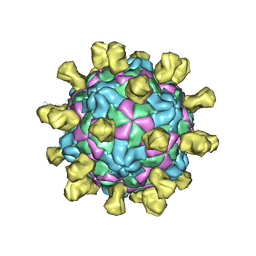 | | Poliovirus (Type 1 Mahoney), receptor-catalysed 135S particle incubated with anti-VP1 mAb at RT for 1 hr | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
5LIA
 
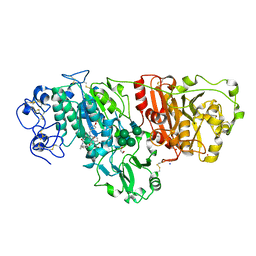 | | Crystal structure of murine autotaxin in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turnbull, A.P, Shah, P, Cheasty, A, Raynham, T, Pang, L, Owen, P. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of potent inhibitors of the lysophospholipase autotaxin.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
1O23
 
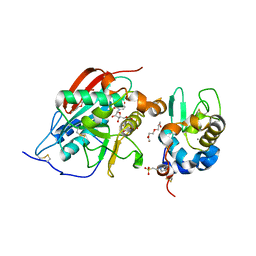 | | CRYSTAL STRUCTURE OF LACTOSE SYNTHASE IN THE PRESENCE OF UDP-GLUCOSE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-01-29 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Alpha-Lactalbumin (La) Stimulates Milk Beta-1,4-Galactosyltransferase I (Beta 4Gal-T1) to Transfer Glucose from Udp-Glucose to N-Acetylglucosamine. Crystal Structure of Beta 4Gal-T1 X La Complex with Udp-Glc.
J.Biol.Chem., 276, 2001
|
|
1NWG
 
 | | BETA-1,4-GALACTOSYLTRANSFERASE COMPLEX WITH ALPHA-LACTALBUMIN AND N-BUTANOYL-GLUCOAMINE | | Descriptor: | 2-(butanoylamino)-2-deoxy-beta-D-glucopyranose, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-02-06 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | ALPHA-LACTALBUMIN (LA) STIMULATES MILK
BETA-1,4-GALACTOSYLTRANSFERASE I (BETA 4GAL-T1) TO
TRANSFER GLUCOSE FROM UDP-GLUCOSE TO
N-ACETYLGLUCOSAMINE. CRYSTAL STRUCTURE OF BETA
4GAL-T1 X LA COMPLEX WITH UDP-GLC.
J.Biol.Chem., 276, 2001
|
|
1NMM
 
 | | beta-1,4-galactosyltransferase mutant Cys342Thr complex with alpha-lactalbumin and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | alpha-Lactalbumin (LA) stimulates milk beta-1,4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine. Crystal structure of beta 4Gal-T1 x LA complex with UDP-Glc.
J.Biol.Chem., 276, 2001
|
|
1ROV
 
 | | Lipoxygenase-3 Treated with Cumene Hydroperoxide | | Descriptor: | FE (III) ION, Seed lipoxygenase-3 | | Authors: | Vahedi-Faridi, A, Brault, P.A, Shah, P, Kim, Y.W, Dunham, W.R, Funk, M.O. | | Deposit date: | 2003-12-02 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction between non-heme iron of lipoxygenases and cumene hydroperoxide: basis for enzyme activation, inactivation, and inhibition
J.Am.Chem.Soc., 126, 2004
|
|
