2LNA
 
 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
1BNL
 
 | | ZINC DEPENDENT DIMERS OBSERVED IN CRYSTALS OF HUMAN ENDOSTATIN | | Descriptor: | COLLAGEN XVIII, ZINC ION | | Authors: | Ding, Y.-H, Javaherian, K, Lo, K.-M, Chopra, R, Boehm, T, Lanciotti, J, Harris, B.A, Li, Y, Shapiro, R, Hohenester, E, Timpl, R, Folkman, J, Wiley, D.C. | | Deposit date: | 1998-07-30 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zinc-dependent dimers observed in crystals of human endostatin.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3LRQ
 
 | | Crystal structure of the U-box domain of human ubiquitin-protein ligase (E3), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D. | | Descriptor: | E3 ubiquitin-protein ligase TRIM37, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | E3 UBIQUITIN-PROTEIN LIGASE RING DOMAIN FROM HUMAN TRIPARTITE MOTIF-CONTAINING PROTEIN 37 (TRIM37), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D.
To be Published
|
|
8ILB
 
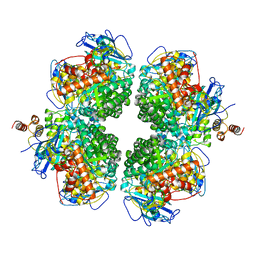 | | The complexes of RbcL, AtRaf1 and AtBSD2 (LFB) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8ILM
 
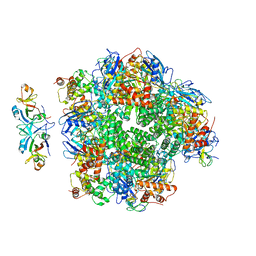 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IO2
 
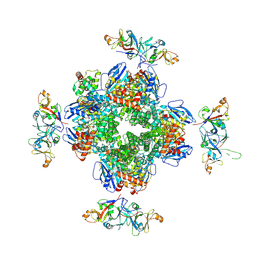 | | The Rubisco assembly intermidate of Arabidopsis thaliana Rubisco accumulation factor 1 (AtRaf1) and Rubisco large subunit (RbcL) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1.2, chloroplastic | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IOJ
 
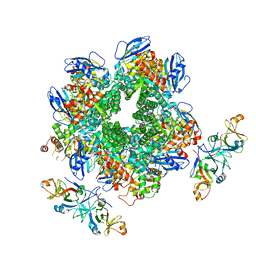 | | The Rubisco assembly intermidiate of Rubisco large subunit (RbcL) and Arabidopsis thaliana Rubisco accumulation factor 1 (AtRaf1) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1.2, chloroplastic | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IOL
 
 | | The complex of Rubisco large subunit (RbcL) | | Descriptor: | Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
1YNY
 
 | | Molecular Structure of D-Hydantoinase from a Bacillus sp. AR9: Evidence for mercury inhibition | | Descriptor: | D-Hydantoinase, MANGANESE (II) ION | | Authors: | Radha Kishan, K.V, Vohra, R.M, Ganeshan, K, Agrawal, V, Sharma, V.M, Sharma, R. | | Deposit date: | 2005-01-26 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of D-hydantoinase from Bacillus sp. AR9: evidence for mercury inhibition.
J.Mol.Biol., 347, 2005
|
|
3PUO
 
 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
3PS7
 
 | | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Gautam, A, Kumar, S, Singh, A, Singh, N, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
3NOE
 
 | | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Kumar, S, Singh, N, Gautam, A, Sharma, R, Sharma, S, Tewari, R, Singh, T.P. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa
To be Published
|
|
1ANG
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN REVEALS THE STRUCTURAL BASIS FOR ITS FUNCTIONAL DIVERGENCE FROM RIBONUCLEASE | | Descriptor: | ANGIOGENIN | | Authors: | Acharya, K.R, Allen, S, Shapiro, R, Riordan, J.F, Vallee, B.L. | | Deposit date: | 1994-01-18 | | Release date: | 1995-04-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJT
 
 | | Ternary complex of E.coli leucyl-tRNA synthetase, tRNA(Leu)574 and the benzoxaborole AN3017 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJU
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
8E20
 
 | |
8EDM
 
 | |
4A7G
 
 | | Structure of human I113T SOD1 mutant complexed with 4-methylpiperazin- 1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)QUINAZOLINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Sharma, R, Antonyuk, S.V, Strange, R.W, Berry, N.G, O'Neil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Curr.Med.Chem., 20, 2013
|
|
1AGI
 
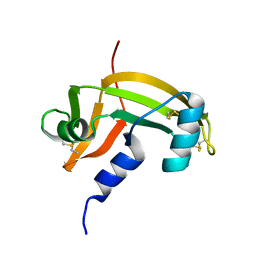 | |
2DO8
 
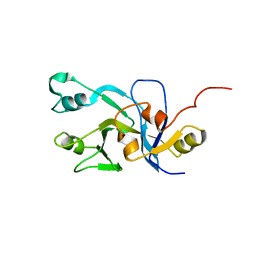 | | Solution Structure of UPF0301 protein HD_1794 | | Descriptor: | UPF0301 protein HD_1794 | | Authors: | Zhang, Q, Liu, G, Shastry, R, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T.R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of UPF0301 protein HD_1794
To be published
|
|
1E94
 
 | | HslV-HslU from E.coli | | Descriptor: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | Deposit date: | 2000-10-07 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6NAZ
 
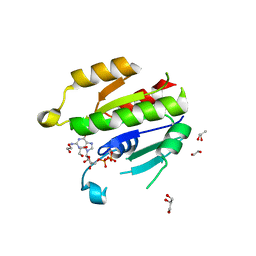 | | Crystal structure of DIRAS 2/3 chimera in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GTP-binding protein Di-Ras2,GTP-binding protein Di-Ras3,GTP-binding protein Di-Ras2, ... | | Authors: | Gilbert, Y.H, Reger, A, Sharma, R, Bast, J, Kim, C. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Crystal structure of DIRAS 2/3 chimera in complex with GDP
To Be Published
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
