4XCK
 
 | | Vibrio cholerae O395 Ribokinase complexed with ADP, Ribose and Cesium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CESIUM ION, Ribokinase, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation.
Adv. Exp. Med. Biol., 842, 2015
|
|
4UXD
 
 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
1F51
 
 | | A TRANSIENT INTERACTION BETWEEN TWO PHOSPHORELAY PROTEINS TRAPPED IN A CRYSTAL LATTICE REVEALS THE MECHANISM OF MOLECULAR RECOGNITION AND PHOSPHOTRANSFER IN SINGAL TRANSDUCTION | | Descriptor: | MAGNESIUM ION, SPORULATION INITIATION PHOSPHOTRANSFERASE B, SPORULATION INITIATION PHOSPHOTRANSFERASE F | | Authors: | Zapf, J, Sen, U, Madhusudan, M, Hoch, J.A, Varughese, K.I. | | Deposit date: | 2000-06-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction.
Structure Fold.Des., 8, 2000
|
|
4QG5
 
 | |
7V4E
 
 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, VpsR | | Authors: | Chakrabortty, T, Sen, U. | | Deposit date: | 2021-08-12 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
7V3W
 
 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VpsR | | Authors: | Chakrabortty, T, Sen, U, Chowdhury, S.R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
7V2V
 
 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | SULFATE ION, VpsR | | Authors: | Chakrabortty, T, Sen, U, Chowdhury, S.R. | | Deposit date: | 2021-08-10 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
7V2B
 
 | |
3TO5
 
 | |
2ET2
 
 | | Crystal structure of an Asn to Ala mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2ESU
 
 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
3I29
 
 | | Crystal structure of a binary complex between an mutant trypsin inhibitor with bovine trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Chymotrypsin inhibitor 3 | | Authors: | Khamrui, S, Majumder, S, Dasgupta, J, Dattagupta, J.K, Sen, U. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Role of remote scaffolding residues in the inhibitory loop pre-organization, flexibility, rigidification and enzyme inhibition of serine protease inhibitors
Biochim.Biophys.Acta, 1824, 2012
|
|
3H2X
 
 | |
3IQS
 
 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
1PEY
 
 | | Crystal structure of the Response Regulator Spo0F complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Sporulation initiation phosphotransferase F | | Authors: | Mukhopadhyay, D, Sen, U, Zapf, J, Varughese, K.I. | | Deposit date: | 2003-05-22 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metals in the sporulation phosphorelay: manganese binding by the response regulator Spo0F.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WBC
 
 | | CRYSTALLIZATION AND PRELIMINARY X-RAY STUDIES OF PSOPHOCARPIN B1, A CHYMOTRYPSIN INHIBITOR FROM WINGED BEAN SEEDS | | Descriptor: | CHYMOTRYPSIN INHIBITOR (WCI) | | Authors: | Dattagupta, J.K, Podder, A, Chakrabarti, C, Sen, U, Dutta, S.K, Singh, M. | | Deposit date: | 1995-11-30 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Kunitz-type chymotrypsin from winged bean seeds at 2.95 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2BEA
 
 | | Crystal structure of Asn14 to Gly mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2BEB
 
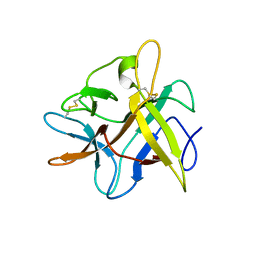 | | X-ray structure of Asn to Thr mutant of Winged Bean Chymotrypsin inhibitor | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
3E1U
 
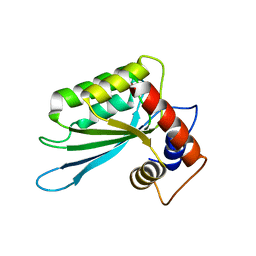 | | The Crystal Structure of the Anti-Viral APOBEC3G Catalytic Domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2008-08-04 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
2QYI
 
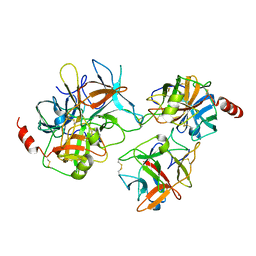 | | Crystal structure of a binary complex between an engineered trypsin inhibitor and Bovine trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Chymotrypsin inhibitor 3, ... | | Authors: | Khamrui, S, Dasgupta, J, Dattagupta, J.K, Sen, U. | | Deposit date: | 2007-08-15 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a binary complex between an engineered trypsin inhibitor and Bovine trypsin
To be Published
|
|
2WBC
 
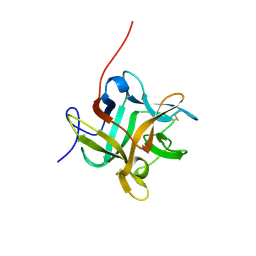 | | REFINED CRYSTAL STRUCTURE (2.3 ANGSTROM) OF A WINGED BEAN CHYMOTRYPSIN INHIBITOR AND LOCATION OF ITS SECOND REACTIVE SITE | | Descriptor: | CHYMOTRYPSIN INHIBITOR | | Authors: | Dattagupta, J.K, Podder, A, Chakrabarti, C, Sen, U, Mukhopadhyay, D, Dutta, S.K, Singh, M. | | Deposit date: | 1997-11-26 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structure (2.3 A) of a double-headed winged bean alpha-chymotrypsin inhibitor and location of its second reactive site.
Proteins, 35, 1999
|
|
1NQP
 
 | | Crystal structure of Human hemoglobin E at 1.73 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Dasgupta, J, Sen, U, Choudhury, D, Dutta, P, Basu, S, Chakrabarti, A, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2003-01-22 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallization and preliminary X-ray structural Studies of Hemoglobin A2 and Hemoglobin E, isolated from the blood samples of Beta-thalassemic patients
Biochem.Biophys.Res.Commun., 303, 2004
|
|
3JQZ
 
 | | Crystal Structure of Human serum albumin complexed with Lidocaine | | Descriptor: | 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, Serum albumin | | Authors: | Hein, K.L, Kragh-Hansen, U, Morth, J.P, Nissen, P. | | Deposit date: | 2009-09-08 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin.
J.Struct.Biol., 2010
|
|
3JRY
 
 | | Human Serum albumin with bound Sulfate | | Descriptor: | SULFATE ION, Serum albumin | | Authors: | Hein, K.L, Kragh-Hansen, U, Morth, J.P, Nissen, P. | | Deposit date: | 2009-09-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin.
J.Struct.Biol., 171, 2010
|
|
6S7C
 
 | | Crystal structure of CARM1 in complex with inhibitor UM079 | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]propyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
