1Z1P
 
 | |
1XPK
 
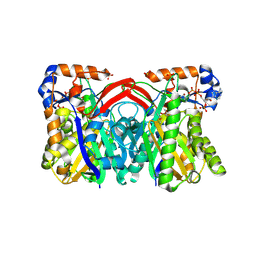 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCUS AUREUS HMG-COA SYNTHASE WITH HMG-CoA AND WITH ACETOACETYL-COA AND ACETYLATED CYSTEINE | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, 3-hydroxy-3-methylglutaryl CoA synthase, ACETOACETYL-COENZYME A, ... | | Authors: | Theisen, M.J, Misra, I, Saadat, D, Campobasso, N, Miziorko, H.M, Harrison, D.H.T. | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in "real-time"
Proc.Natl.Acad.Sci.USA, 47, 2004
|
|
3D77
 
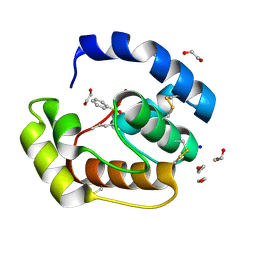 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 4.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
1Z1Q
 
 | |
2A50
 
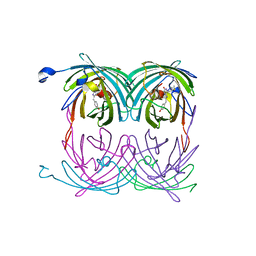 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UUO
 
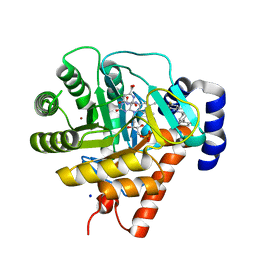 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with brequinar | | Descriptor: | 6-FLUORO-2-(2'-FLUORO-1,1'-BIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
2A54
 
 | | fluorescent protein asFP595, A143S, on-state, 1min irradiation | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
287D
 
 | |
2A52
 
 | | fluorescent protein asFP595, S158V, on-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1SIZ
 
 | |
1SJ1
 
 | | The 1.5 A Resolution Crystal Structure of [Fe3S4]-Ferredoxin from the hyperthermophilic Archaeon Pyrococcus furiosus | | Descriptor: | COBALT HEXAMMINE(III), FE3-S4 CLUSTER, Ferredoxin | | Authors: | Nielsen, M.S, Harris, P, Ooi, B.L, Christensen, H.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A Resolution Crystal Structure of [Fe3S4]-Ferredoxin from the Hyperthermophilic Archaeon Pyrococcus furiosus
Biochemistry, 43, 2004
|
|
2A53
 
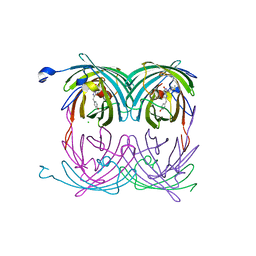 | | fluorescent protein asFP595, A143S, off-state | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4AKD
 
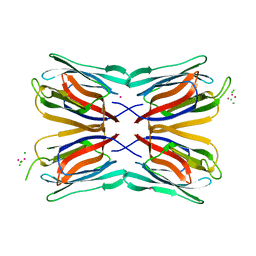 | | High resolution structure of Mannose Binding lectin from Champedak (CMB) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MANNOSE-SPECIFIC LECTIN KM+ | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
2A56
 
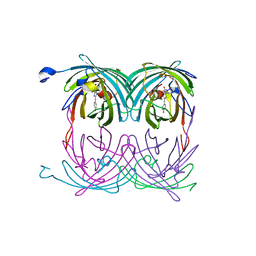 | | fluorescent protein asFP595, A143S, on-state, 5min irradiation | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2K37
 
 | | CsmA | | Descriptor: | Chlorosome Protein A | | Authors: | Pedersen, M, Dittmer, J, Underhaug, J, Miller, M, Nielsen, N.C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of CsmA: A small antenna protein from the green sulfur bacterium Chlorobium tepidum.
Febs Lett., 582, 2008
|
|
1S46
 
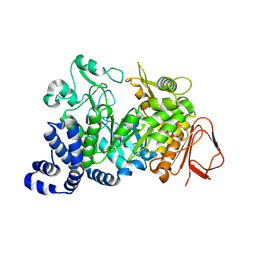 | | Covalent intermediate of the E328Q amylosucrase mutant | | Descriptor: | amylosucrase, beta-D-glucopyranose | | Authors: | Jensen, M.H, Mirza, O, Albenne, C, Remaud-Simeon, M, Monsan, P, Gajhede, M, Skov, L.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea.
Biochemistry, 43, 2004
|
|
1KUN
 
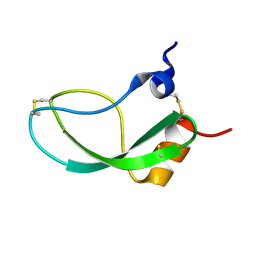 | | SOLUTION STRUCTURE OF THE HUMAN ALPHA3-CHAIN TYPE VI COLLAGEN C-TERMINAL KUNITZ DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | ALPHA3-CHAIN TYPE VI COLLAGEN | | Authors: | Sorensen, M.D, Bjorn, S, Norris, K, Olsen, O, Petersen, L, James, T.L, Led, J.J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the human alpha3-chain type VI collagen C-terminal Kunitz domain,.
Biochemistry, 36, 1997
|
|
1AJW
 
 | |
4AKB
 
 | | Structure of Galactose Binding lectin from Champedak (CGB) with Galactose | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA-4 CHAIN, HEXAETHYLENE GLYCOL, ... | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4AKC
 
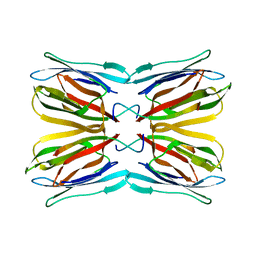 | | Structure of Galactose Binding lectin from Champedak (CGB) with Gal(beta)1,3-GalNac | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA-4 CHAIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1NHY
 
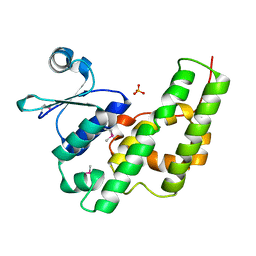 | | Crystal Structure of the GST-like Domain of Elongation Factor 1-gamma from Saccharomyces cerevisiae. | | Descriptor: | Elongation factor 1-gamma 1, SULFATE ION | | Authors: | Jeppesen, M.G, Ortiz, P, Kinzy, T.G, Andersen, G.R, Nyborg, J. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Glutathione S-Transferase-like Domain of Elongation Factor 1B{gamma} from Saccharomyces cerevisiae.
J.Biol.Chem., 278, 2003
|
|
189D
 
 | |
187D
 
 | |
1S6Z
 
 | | Enhanced Green Fluorescent Protein Containing the Y66L Substitution | | Descriptor: | CHLORIDE ION, green fluorescent protein | | Authors: | Rosenow, M.A, Huffman, H.A, Phail, M.E, Wachter, R.M. | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Y66L Variant of Green Fluorescent Protein Supports a Cyclization-Oxidation-Dehydration Mechanism for Chromophore Maturation(,).
Biochemistry, 43, 2004
|
|
1O73
 
 | | Tryparedoxin from Trypanosoma brucei | | Descriptor: | TRYPAREDOXIN | | Authors: | Gabrielsen, M, Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-10-23 | | Release date: | 2003-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
