5RUB
 
 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE FROM RHODOSPIRILLUM RUBRUM AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Schneider, G, Lindqvist, Y, Lundqvist, T. | | Deposit date: | 1990-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution.
J.Mol.Biol., 211, 1990
|
|
6HNG
 
 | |
6HNE
 
 | |
1TKB
 
 | |
1TKA
 
 | |
5MXL
 
 | |
5MXT
 
 | |
5OJH
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRATE ANION, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Schneider, G, Vella, P, Lindqvist, Y, Schnell, R. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
1TKC
 
 | |
4IP2
 
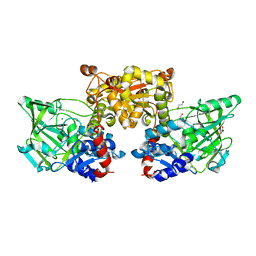 | | Putative Aromatic Acid Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, Aromatic Acid Decarboxylase, GLYCEROL, ... | | Authors: | Schneider, G, Brunner, K, Izumi, A, Jacewicz, A. | | Deposit date: | 2013-01-09 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the UbiD protein family from the crystal structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
3EUL
 
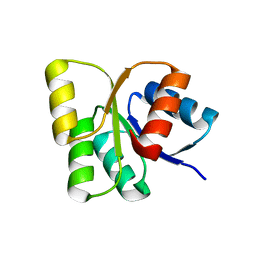 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3R8R
 
 | |
1RBA
 
 | |
6HNH
 
 | |
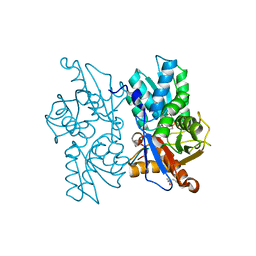
2Q3D
 
 | |
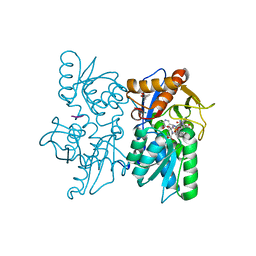
2Q3C
 
 | |
2Q3B
 
 | |
1F05
 
 | |
9RUB
 
 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | Descriptor: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1990-11-28 | | Release date: | 1993-01-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
8PBO
 
 | | Deep interactome learning for generative drug design | | Descriptor: | 3-[2-fluoranyl-4-[3-[2-fluoranyl-4-(5-methyl-1,3,4-thiadiazol-2-yl)phenoxy]propoxy]phenyl]propanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Hakansson, M, Focht, D, Atz, K, Schneider, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Prospective de novo drug design with deep interactome learning.
Nat Commun, 15, 2024
|
|
7QZJ
 
 | |
5A6O
 
 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A6N
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 | | Descriptor: | 5-(3-SULFAMOYLPHENYL)-1H-1,2,3,4-TETRAZOL-1-IDE, DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, ... | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2Y5T
 
 | | Crystal structure of the pathogenic autoantibody CIIC1 in complex with the triple-helical C1 peptide | | Descriptor: | C1, CHLORIDE ION, CIIC1 FAB FRAGMENT HEAVY CHAIN, ... | | Authors: | Dobritzsch, D, Lindh, I, Schneider, N, Uysal, H, Nandakumar, K.S, Burkhardt, H, Schneider, G, Holmdahl, R. | | Deposit date: | 2011-01-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Arthritis Rheum., 63, 2011
|
|
4S2B
 
 | | Covalent complex of E. coli transaldolase TalB with tagatose-6-phosphate | | Descriptor: | 2-deoxy-6-O-phosphono-beta-D-lyxo-hexofuranose, SULFATE ION, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
