7QZJ
 
 | |
6YKD
 
 | | Human Pim-1 kinase in complex with an inhibitor identified by virtual screening | | Descriptor: | ACETATE ION, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Schneider, P, Welin, M, Svensson, B, Walse, B, Schneider, G. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Virtual Screening and Design with Machine Intelligence Applied to Pim-1 Kinase Inhibitors.
Mol Inform, 39, 2020
|
|
9RUB
 
 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | Descriptor: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1990-11-28 | | Release date: | 1993-01-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
1QPB
 
 | | PYRUVATE DECARBOYXLASE FROM YEAST (FORM B) COMPLEXED WITH PYRUVAMIDE | | Descriptor: | MAGNESIUM ION, PYRUVAMIDE, PYRUVATE DECARBOXYLASE (FORM B), ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1999-11-26 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis of Substrate Activation in Yeast Pyruvate Decarboxylase a Crystallographic and Kinetic Study
Eur.J.Biochem., 267, 2000
|
|
5A6N
 
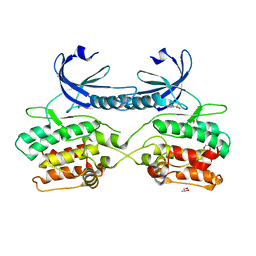 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 | | Descriptor: | 5-(3-SULFAMOYLPHENYL)-1H-1,2,3,4-TETRAZOL-1-IDE, DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, ... | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A6O
 
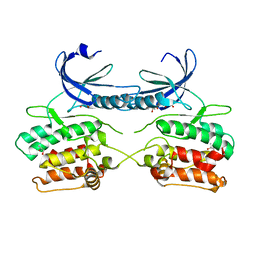 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1GT8
 
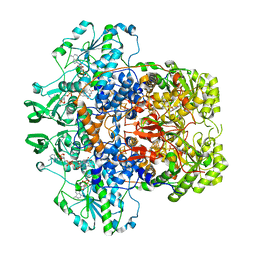 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTH
 
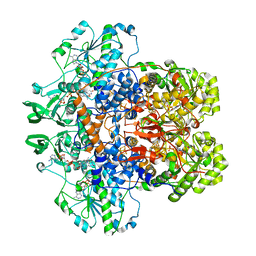 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
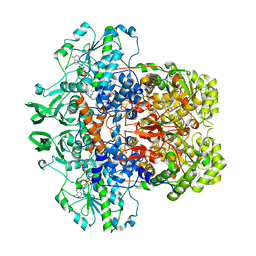 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1QZZ
 
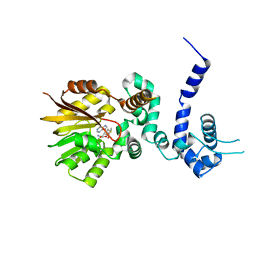 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1R00
 
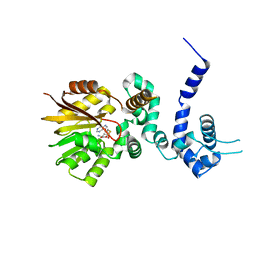 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-homocysteine (SAH) | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G. | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
4AG3
 
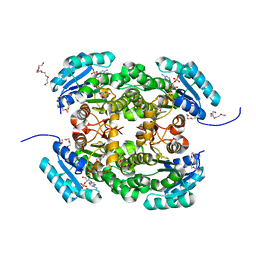 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with NADPH at 1.8A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | Authors: | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | Deposit date: | 2012-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
2Q0K
 
 | | Oxidized thioredoxin reductase from Helicobacter pylori in complex with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase | | Authors: | Sandalova, T, Gustafsson, T, Lu, J, Holmgren, A, Schneider, G. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1QHW
 
 | | PURPLE ACID PHOSPHATASE FROM RAT BONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PROTEIN (PURPLE ACID PHOSPHATASE), ... | | Authors: | Lindqvist, Y, Johansson, E, Kaija, H, Vihko, P, Schneider, G. | | Deposit date: | 1999-03-26 | | Release date: | 1999-09-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a mammalian purple acid phosphatase at 2.2 A resolution with a mu-(hydr)oxo bridged di-iron center.
J.Mol.Biol., 291, 1999
|
|
2Q0L
 
 | | Helicobacter pylori thioredoxin reductase reduced by sodium dithionite in complex with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase | | Authors: | Sandalova, T, Gustafsson, T, Lu, J, Holmgren, A, Schneider, G. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QA1
 
 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
8PBO
 
 | | Deep interactome learning for generative drug design | | Descriptor: | 3-[2-fluoranyl-4-[3-[2-fluoranyl-4-(5-methyl-1,3,4-thiadiazol-2-yl)phenoxy]propoxy]phenyl]propanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Hakansson, M, Focht, D, Atz, K, Schneider, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Prospective de novo drug design with deep interactome learning.
Nat Commun, 15, 2024
|
|
1QJ3
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase in complex with 7-keto-8-aminopelargonic acid | | Descriptor: | 7,8-DIAMINOPELARGONIC ACID SYNTHASE, 7-KETO-8-AMINOPELARGONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Diaminopelargonic Acid Synthase; Evolutionary Relationships between Pyridoxal-5'-Phosphate Dependent Enzymes
J.Mol.Biol., 291, 1999
|
|
1QJ5
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase | | Descriptor: | 7,8-DIAMINOPELARGONIC ACID SYNTHASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kack, H, Sandmark, J, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Diaminopelargonic Acid Synthase; Evolutionary Relationships between Pyridoxal-5'-Phosphate Dependent Enzymes
J.Mol.Biol., 291, 1999
|
|
1R0K
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Zymomonas mobilis | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ACETATE ION | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1R0L
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from zymomonas mobilis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
4S2C
 
 | | Covalent complex of E. coli transaldolase TalB with fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, FRUCTOSE -6-PHOSPHATE, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4S2B
 
 | | Covalent complex of E. coli transaldolase TalB with tagatose-6-phosphate | | Descriptor: | 2-deoxy-6-O-phosphono-beta-D-lyxo-hexofuranose, SULFATE ION, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2Y5T
 
 | | Crystal structure of the pathogenic autoantibody CIIC1 in complex with the triple-helical C1 peptide | | Descriptor: | C1, CHLORIDE ION, CIIC1 FAB FRAGMENT HEAVY CHAIN, ... | | Authors: | Dobritzsch, D, Lindh, I, Schneider, N, Uysal, H, Nandakumar, K.S, Burkhardt, H, Schneider, G, Holmdahl, R. | | Deposit date: | 2011-01-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Arthritis Rheum., 63, 2011
|
|
2QA2
 
 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
