1PPZ
 
 | | Trypsin complexes at atomic and ultra-high resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
6G1Y
 
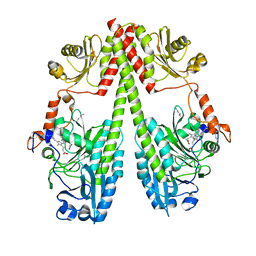 | | Crystal structure of the photosensory core module (PCM) of a bathy phytochrome from Agrobacterium fabrum in the Pfr state. | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Schmidt, A, Qureshi, B.M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
1IS9
 
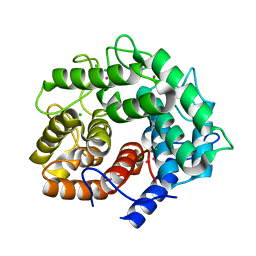 | | Endoglucanase A from Clostridium thermocellum at atomic resolution | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, endoglucanase A | | Authors: | Schmidt, A, Gonzalez, A, Morris, R.J, Costabel, M, Alzari, P.M, Lamzin, V.S. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Advantages of high-resolution phasing: MAD to atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8POU
 
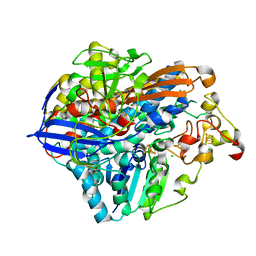 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.65 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POV
 
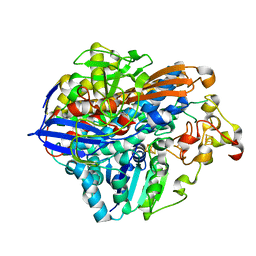 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.92 A Resolution. | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
7ODH
 
 | |
7ODG
 
 | |
5MDJ
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in a its as-isolated high-pressurized form | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MDK
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form (oxidized state - state 3) | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4HKB
 
 | | CH67 Fab (unbound) from the CH65-67 Lineage | | Descriptor: | CH67 heavy chain, CH67 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK3
 
 | | I2 Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HKX
 
 | | Influenza hemagglutinin in complex with CH67 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH67 heavy chain, CH67 light chain, ... | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK0
 
 | | UCA Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1XVO
 
 | | Trypsin from Fusarium oxysporum at pH 6 | | Descriptor: | SULFATE ION, trypsin | | Authors: | Schmidt, A, Lamzin, V.S. | | Deposit date: | 2004-10-28 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Extraction of functional motion in trypsin crystal structures.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1FC4
 
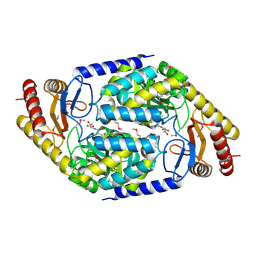 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1DIB
 
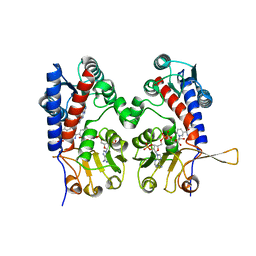 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
4HTW
 
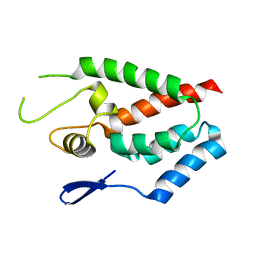 | | SIVmac239 capsid N-terminal domain | | Descriptor: | Gag protein | | Authors: | Schmidt, A.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gain-of-Sensitivity Mutations in a Trim5-Resistant Primary Isolate of Pathogenic SIV Identify Two Independent Conserved Determinants of Trim5alpha Specificity.
Plos Pathog., 9, 2013
|
|
1XVM
 
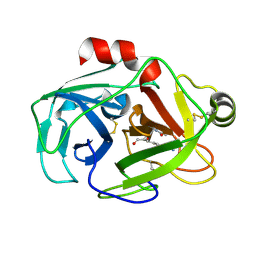 | |
1DIG
 
 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY374571 | | Descriptor: | ACETATE ION, METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIA
 
 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY249543 | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [[[2-AMINO-5,6,7,8-TETRAHYDRO-4-HYDROXY-PYRIDO[2,3-D]PYRIMIDIN-6-YL]-ETHYL]-PHENYL]-CARBONYL-GLUTAMIC ACID | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1ZR0
 
 | | Crystal Structure of Kunitz Domain 1 of Tissue Factor Pathway Inhibitor-2 with Bovine Trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Tissue factor pathway inhibitor 2 | | Authors: | Schmidt, A.E, Chand, H.S, Cascio, D, Kisiel, W, Bajaj, S.P. | | Deposit date: | 2005-05-18 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of Kunitz domain 1 (KD1) of tissue factor pathway inhibitor-2 in complex with trypsin. Implications for KD1 specificity of inhibition
J.Biol.Chem., 280, 2005
|
|
4TTT
 
 | |
2I9E
 
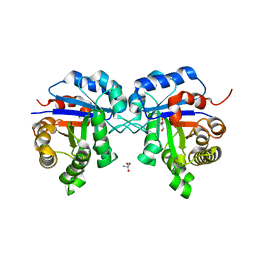 | | Structure of Triosephosphate Isomerase of Tenebrio molitor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Triosephosphate isomerase | | Authors: | Schmidt, A, Scheerer, P, Wessner, H, Hoehne, W, Krauss, N. | | Deposit date: | 2006-09-05 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A coleopteran triosephosphate isomerase: X-ray structure and phylogenetic impact of insect sequences.
Insect Mol Biol, 19, 2010
|
|
4V4E
 
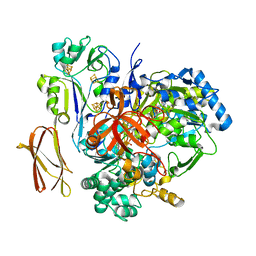 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4V4C
 
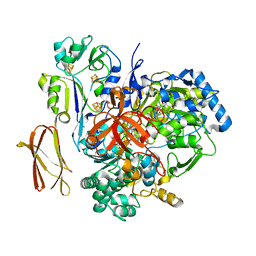 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
