3GMP
 
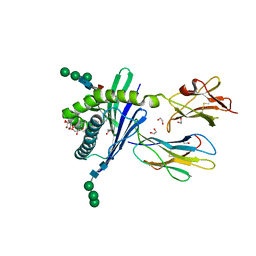 | | Structure of mouse CD1d in complex with PBS-25 | | Descriptor: | (2S,3S,4R)-N-OCTANOYL-1-[(ALPHA-D-GALACTOPYRANOSYL)OXY]-2-AMINO-OCTADECANE-3,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMN
 
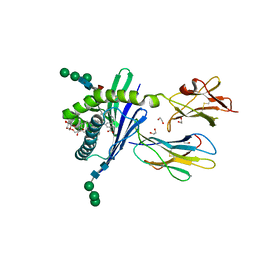 | | Structure of mouse CD1d in complex with C10Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
4GH7
 
 | |
1SW2
 
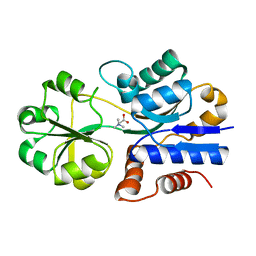 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with glycine betaine | | Descriptor: | TRIMETHYL GLYCINE, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1SW5
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in the ligand free form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1SW4
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with trimethyl ammonium | | Descriptor: | CHLORIDE ION, TETRAMETHYLAMMONIUM ION, ZINC ION, ... | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
4KRX
 
 | | Structure of Aes from E. coli | | Descriptor: | Acetyl esterase, TETRAETHYLENE GLYCOL | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
1SW1
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, ZINC ION, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
4IDC
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADPH and HDMF | | Descriptor: | (2R)-4-hydroxy-2,5-dimethylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
4IDB
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Ripening-induced protein, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
4IDD
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADPH and EHMF | | Descriptor: | (2R)-2-ethyl-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
4IDE
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADP+ and EDHMF | | Descriptor: | (2E)-2-ethylidene-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
4IDA
 
 | |
4IDF
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADPH and HMF | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-5-methylfuran-3(2H)-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
4KRY
 
 | | Structure of Aes from E. coli in covalent complex with PMS | | Descriptor: | Acetyl esterase, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
3KQ0
 
 | | Crystal structure of human alpha1-acid glycoprotein | | Descriptor: | (2R)-2,3-dihydroxypropyl acetate, Alpha-1-acid glycoprotein 1, CHLORIDE ION | | Authors: | Schiefner, A, Schonfeld, D.L, Ravelli, R.B.G, Mueller, U, Skerra, A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8-A crystal structure of alpha1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin.
J.Mol.Biol., 384, 2008
|
|
3CJJ
 
 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
2QSC
 
 | | Crystal structure analysis of anti-HIV-1 V3-Fab F425-B4e8 in complex with a V3-peptide | | Descriptor: | CHLORIDE ION, Envelope glycoprotein gp120, Fab F425-B4e8, ... | | Authors: | Bell, C.H, Schiefner, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization.
J.Mol.Biol., 375, 2008
|
|
4S3P
 
 | |
4S3R
 
 | |
2GIF
 
 | | Asymmetric structure of trimeric AcrB from Escherichia coli | | Descriptor: | Acriflavine resistance protein B, CITRATE ANION | | Authors: | Seeger, M.A, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K, Pos, K.M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism.
Science, 313, 2006
|
|
4S3Q
 
 | | Amylomaltase MalQ from Escherichia coli in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase, IODIDE ION, ... | | Authors: | Weiss, S.C, Schiefner, A. | | Deposit date: | 2015-03-26 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Interconversion of Maltodextrins by MalQ, the Amylomaltase of Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
7O31
 
 | | Crystal structure of the anti-PAS Fab 1.2 in complex with its epitope peptide and the anti-Kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, PAS#1 epitope peptide, anti-Kappa VHH domain, ... | | Authors: | Schilz, J, Schiefner, A, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
7O2Z
 
 | | Crystal structure of the anti-PAS Fab 2.2 in complex with its epitope peptide | | Descriptor: | CHLORIDE ION, P/A#1 epitope peptide, anti-PAS Fab 2.2 chimeric heavy chain, ... | | Authors: | Schilz, J, Schiefner, A, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
7O30
 
 | |
