1KY2
 
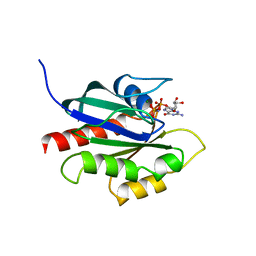 | | GPPNHP-BOUND YPT7P AT 1.6 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1XNJ
 
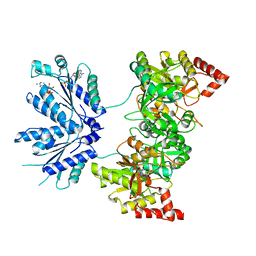 | | APS complex of human PAPS synthetase 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 | | Authors: | Harjes, S, Bayer, P, Scheidig, A.J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of human PAPS synthetase 1 reveals asymmetry in substrate binding
J.Mol.Biol., 347, 2005
|
|
1KY3
 
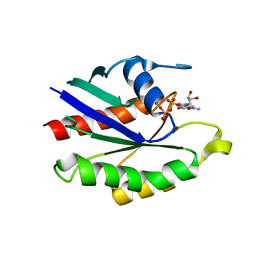 | | GDP-BOUND YPT7P AT 1.35 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1JLN
 
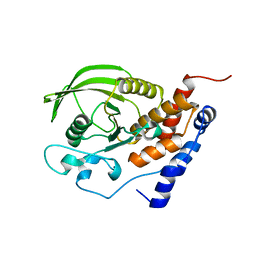 | | Crystal structure of the catalytic domain of protein tyrosine phosphatase PTP-SL/BR7 | | Descriptor: | Protein Tyrosine Phosphatase, receptor type, R | | Authors: | Szedlacsek, S.E, Aricescu, A.R, Fulga, T.A, Renault, L, Scheidig, A.J. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of PTP-SL/PTPBR7 catalytic domain: implications for MAP kinase regulation.
J.Mol.Biol., 311, 2001
|
|
2Z2C
 
 | | MURA inhibited by unag-cnicin adduct | | Descriptor: | 2-acetamido-3-O-[(2S,3S)-2-carboxy-3,4-dihydroxybutan-2-yl]-2-deoxy-alpha-D-glucopyranose, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Steinbach, A, Skarzynski, T, Scheidig, A.J, Klein, C.D. | | Deposit date: | 2007-05-17 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Antibacterial Target Enzyme Mura Synthesizes its Own Non-Covalent Suicide Inhibitor from the Natural Product Cnicin
To be Published
|
|
2WSB
 
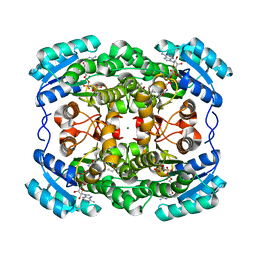 | | Crystal structure of the short-chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD | | Descriptor: | GALACTITOL DEHYDROGENASE, MAGNESIUM ION, N-PROPANOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-09-04 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
2XTS
 
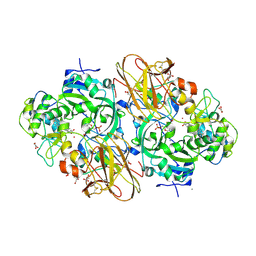 | | Crystal Structure of the Sulfane Dehydrogenase SoxCD from Paracoccus pantotrophus | | Descriptor: | CALCIUM ION, COBALT (II) ION, CYTOCHROME, ... | | Authors: | Zander, U, Faust, A, Klink, B.U, de Sanctis, D, Panjikar, S, Quentmeier, A, Bardischewski, F, Friedrich, C.G, Scheidig, A.J. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
J.Biol.Chem., 286, 2011
|
|
4YAK
 
 | |
4YBZ
 
 | |
4YAJ
 
 | |
4Y8V
 
 | |
4YB8
 
 | |
5LSH
 
 | | human lysozyme in complex with a tetrasaccharide fragment of the O-chain of LPS from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Zhang, R, Nifantiev, N.E, Krylov, V, Luetteke, T, Scheidig, A.J, Siebert, H.-C. | | Deposit date: | 2016-08-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.061 Å) | | Cite: | Lysozyme's lectin-like characteristics facilitates its immune defense function.
Q. Rev. Biophys., 50, 2017
|
|
1AGP
 
 | | THREE-DIMENSIONAL STRUCTURES AND PROPERTIES OF A TRANSFORMING AND A NONTRANSFORMING GLY-12 MUTANT OF P21-H-RAS | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Franken, S.M, Scheidig, A.J, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1993-03-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras.
Biochemistry, 32, 1993
|
|
1CBW
 
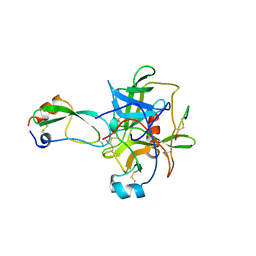 | | BOVINE CHYMOTRYPSIN COMPLEXED TO BPTI | | Descriptor: | BOVINE CHYMOTRYPSIN, BPTI, SULFATE ION | | Authors: | Hynes, T.R, Scheidig, A.J, Kossiakoff, A.A. | | Deposit date: | 1996-12-22 | | Release date: | 1997-07-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities.
Protein Sci., 6, 1997
|
|
2IBT
 
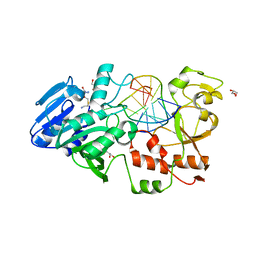 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position and an abasic site analog at the target base partner position | | Descriptor: | 5'-D(*GP*AP*CP*AP*(3DR)P*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
2IH4
 
 | |
2IH5
 
 | |
2IH2
 
 | |
2IBS
 
 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position | | Descriptor: | 5'-D(*GP*AP*CP*AP*TP*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Pljevaljcic, G, Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
2JG3
 
 | | MtaqI with BAZ | | Descriptor: | 5'-D(*GP*AP*CP*AP*TP*CP*GP*6MAP*AP*CP)-3', 5'-D(*GP*TP*TP*CP*GP*AP*TP*GP*TP*CP)-3', 5'-DEOXY-5'-(ETHYLAMINO)-8-{[4-({5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANOYL}AMINO)BUTYL]AMINO}ADENOSINE, ... | | Authors: | Pljevaljcic, G, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2007-02-07 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quantitative Labeling of Long Plasmid DNA with Nanometer Precision.
Chembiochem, 8, 2007
|
|
6S6G
 
 | |
6S49
 
 | |
6S72
 
 | |
6S65
 
 | |
