4U63
 
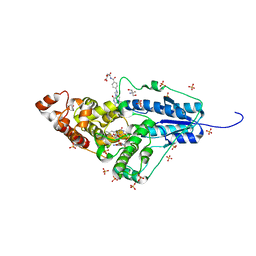 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | 著者 | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | 登録日 | 2014-07-26 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
6R26
 
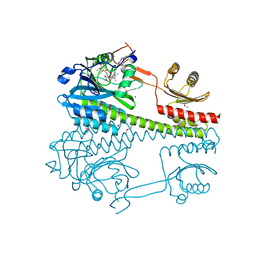 | | The photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | 分子名称: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein, CALCIUM ION | | 著者 | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | 登録日 | 2019-03-15 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R27
 
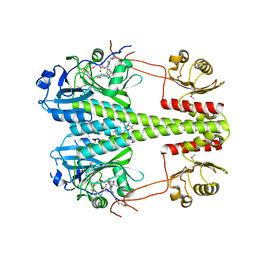 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | 分子名称: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | 著者 | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | 登録日 | 2019-03-15 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
1ZEA
 
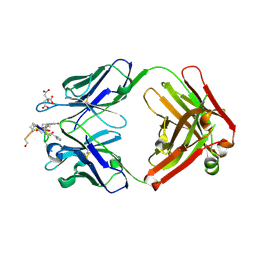 | | Structure of the anti-cholera toxin antibody Fab fragment TE33 in complex with a D-peptide | | 分子名称: | CITRIC ACID, monoclonal anti-cholera toxin IGG1 KAPPA antibody, H chain, ... | | 著者 | Scheerer, P, Krauss, N, Wessner, H, Scholz, C, Otte, L, Seifert, M, Kramer, A, Schneider-Mergener, J, Hoehne, W. | | 登録日 | 2005-04-18 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structure of an anti-cholera toxin antibody Fab in complex with an epitope-derived D-peptide: a case of polyspecific recognition.
J.Mol.Recognit., 20, 2007
|
|
4DJA
 
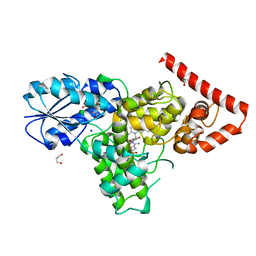 | | Crystal structure of a prokaryotic (6-4) photolyase PhrB from Agrobacterium Tumefaciens with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore at 1.45A resolution | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Scheerer, P, Zhang, F, Oberpichler, I, Lamparter, T, Krauss, N. | | 登録日 | 2012-02-01 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3DQB
 
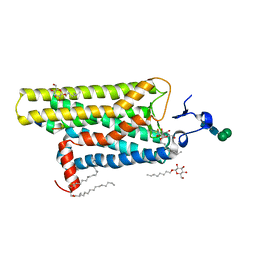 | | Crystal structure of the active G-protein-coupled receptor opsin in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | 分子名称: | 11meric peptide form Guanine nucleotide-binding protein G(t) subunit alpha-1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | 著者 | Scheerer, P, Park, J.H, Hildebrand, P.W, Kim, Y.J, Krauss, N, Choe, H.-W, Hofmann, K.P, Ernst, O.P. | | 登録日 | 2008-07-09 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of opsin in its G-protein-interacting conformation
Nature, 455, 2008
|
|
3ITF
 
 | |
2OBI
 
 | |
3RGW
 
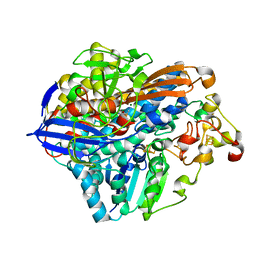 | | Crystal structure at 1.5 A resolution of an H2-reduced, O2-tolerant hydrogenase from Ralstonia eutropha unmasks a novel iron-sulfur cluster | | 分子名称: | FE3-S4 CLUSTER, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Scheerer, P, Fritsch, J, Frielingsdorf, S, Kroschinsky, S, Friedrich, B, Lenz, O, Spahn, C.M.T. | | 登録日 | 2011-04-10 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre.
Nature, 479, 2011
|
|
4V6T
 
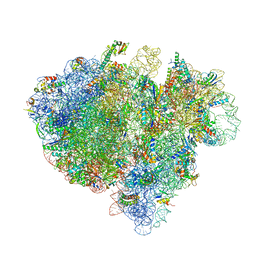 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | 登録日 | 2012-01-27 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.3 Å) | | 主引用文献 | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
8POX
 
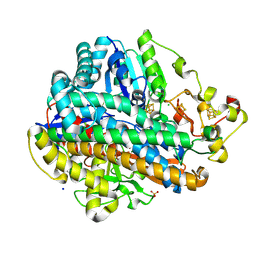 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.6 A Resolution. | | 分子名称: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | 著者 | Kalms, J, Schmidt, A, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POZ
 
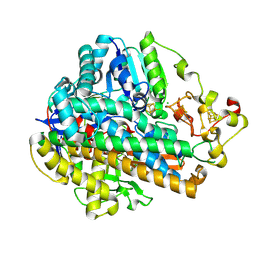 | | Crystal Structure of the C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.65 A Resolution. | | 分子名称: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | 著者 | Schmidt, A, Kalms, J, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2024-09-18 | | 最終更新日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POY
 
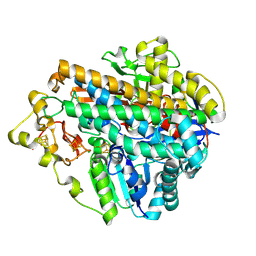 | | Crystal Structure of the C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.93 A Resolution. | | 分子名称: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | 著者 | Schmidt, A, Kalms, J, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
9G8C
 
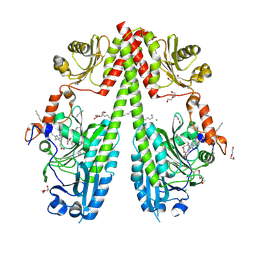 | | Crystal structure of the photosensory core module (PCM) of a cyano-phenylalanine mutant oCNF165 of the bathy phytochrome Agp2 from Agrobacterium fabrum in the Pfr state. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | 著者 | Sauthof, L, Schmidt, A, Scheerer, P. | | 登録日 | 2024-07-23 | | 公開日 | 2024-11-27 | | 最終更新日 | 2024-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Hydrogen Bonding and Noncovalent Electric Field Effects in the Photoconversion of a Phytochrome.
J.Phys.Chem.B, 128, 2024
|
|
9G8D
 
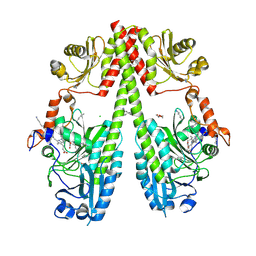 | | Crystal structure of the photosensory core module (PCM) of a cyano-phenylalanine mutant oCNF192 of the bathy phytochrome Agp2 from Agrobacterium fabrum in the Pfr state. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | 著者 | Sauthof, L, Schmidt, A, Scheerer, P. | | 登録日 | 2024-07-23 | | 公開日 | 2024-11-27 | | 最終更新日 | 2024-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.059 Å) | | 主引用文献 | Hydrogen Bonding and Noncovalent Electric Field Effects in the Photoconversion of a Phytochrome.
J.Phys.Chem.B, 128, 2024
|
|
8POU
 
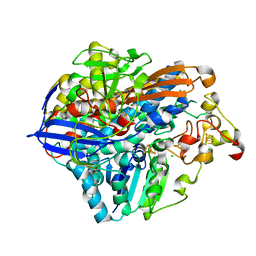 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.65 A Resolution. | | 分子名称: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Schmidt, A, Kalms, J, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POV
 
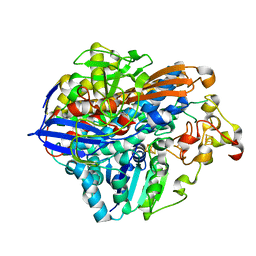 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.92 A Resolution. | | 分子名称: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Schmidt, A, Kalms, J, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POW
 
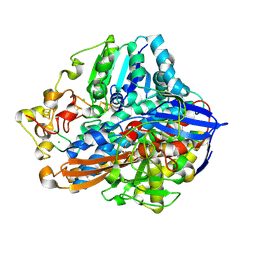 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.61 A Resolution. | | 分子名称: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | 著者 | Kalms, J, Schmidt, A, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | 登録日 | 2015-02-19 | | 公開日 | 2015-05-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
2I9E
 
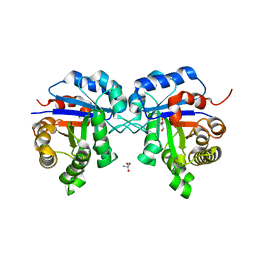 | | Structure of Triosephosphate Isomerase of Tenebrio molitor | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Triosephosphate isomerase | | 著者 | Schmidt, A, Scheerer, P, Wessner, H, Hoehne, W, Krauss, N. | | 登録日 | 2006-09-05 | | 公開日 | 2006-09-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A coleopteran triosephosphate isomerase: X-ray structure and phylogenetic impact of insect sequences.
Insect Mol Biol, 19, 2010
|
|
3CAP
 
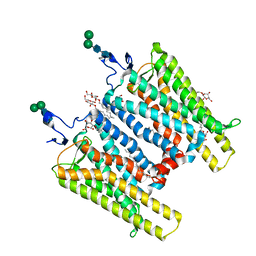 | | Crystal Structure of Native Opsin: the G Protein-Coupled Receptor Rhodopsin in its Ligand-free State | | 分子名称: | 2-O-octyl-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | 著者 | Park, J.H, Scheerer, P, Hofmann, K.P, Choe, H.-W, Ernst, O.P. | | 登録日 | 2008-02-20 | | 公開日 | 2008-06-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the ligand-free G-protein-coupled receptor opsin
Nature, 454, 2008
|
|
7ODH
 
 | |
7ODG
 
 | |
7PIU
 
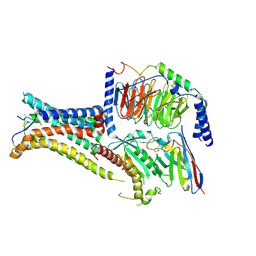 | | Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution. | | 分子名称: | CALCIUM ION, Camelid antibody fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | 登録日 | 2021-08-23 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
7PIV
 
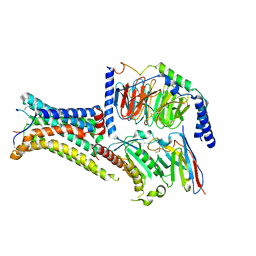 | | Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution. | | 分子名称: | CALCIUM ION, Camelid antibody VHH fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | 登録日 | 2021-08-23 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
