4E0B
 
 | | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6 | | Descriptor: | ACETATE ION, Malate dehydrogenase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-02 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6
To be Published
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7SPN
 
 | | Crystal structure of IS11, a thermophilic esterase | | Descriptor: | IS11 | | Authors: | Stogios, P.J, Evdokimova, E, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of IS11, a thermophilic esterase
To Be Published
|
|
3H2Y
 
 | | Crystal structure of YqeH GTPase from Bacillus anthracis with dGDP bound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, GTPase family protein | | Authors: | Brunzelle, J.S, Anderson, S.M, Xu, X, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of YqeH GTPase from Bacillus anthracis with dGDP Bound
To be Published
|
|
7LAO
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IIb | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside N(3)-acetyltransferase III, MAGNESIUM ION | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
3OT1
 
 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
5KL9
 
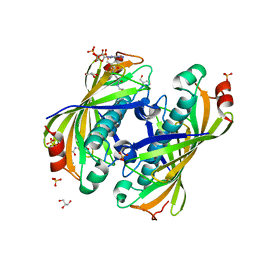 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA
To Be Published
|
|
5KKO
 
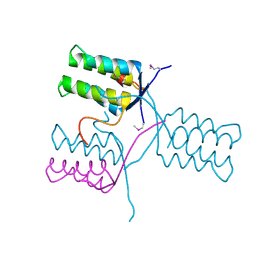 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | Descriptor: | Uncharacterised protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-22 | | Release date: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
7TQ1
 
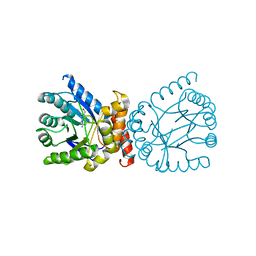 | | Crystal structure of adaptive laboratory evolved sulfonamide-resistant Dihydropteroate Synthase (DHPS) from Escherichia coli in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Dihydropteroate synthase | | Authors: | Stogios, P.J, Skarina, T, Tan, K, Venkatesan, M, Fruci, M, Joachimiak, A, Savchenko, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
6U6A
 
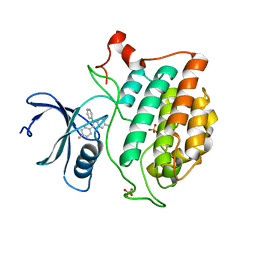 | | Crystal structure of Yck2 from Candida albicans in complex with kinase inhibitor GW461484A | | Descriptor: | 2-(4-fluorophenyl)-6-methyl-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine, SULFATE ION, Serine/threonine protein kinase | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Chang, C, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
3GA7
 
 | | 1.55 Angstrom Crystal Structure of an Acetyl Esterase from Salmonella typhimurium | | Descriptor: | Acetyl esterase, CHLORIDE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J, Onopriyenko, O, Skarina, T, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-16 | | Release date: | 2009-02-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of an Acetyl Esterase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
5L1A
 
 | | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila
To Be Published
|
|
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUM
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with paromomycin and coenzyme A | | Descriptor: | Aminocyclitol acetyltransferase ApmA, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7RH8
 
 | | Crystal structure of Fur1p from Candida albicans, in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, Uracil phosphoribosyltransferase | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Fur1p from Candida albicans, in complex with UTP
To Be Published
|
|
7MX5
 
 | | Crystal structure of TolB from Acinetobacter baumannii | | Descriptor: | Tol-Pal system protein TolB | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of TolB from Acinetobacter baumannii
To Be Published
|
|
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | Descriptor: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
3IAC
 
 | | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium. | | Descriptor: | CHLORIDE ION, Glucuronate isomerase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium.
To be Published
|
|
3IGX
 
 | | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis. | | Descriptor: | PHOSPHATE ION, Transaldolase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis.
TO BE PUBLISHED
|
|
7TBU
 
 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
5JOQ
 
 | | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, CITRIC ACID, Lmo2184 protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-02 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e
To Be Published
|
|
