7M53
 
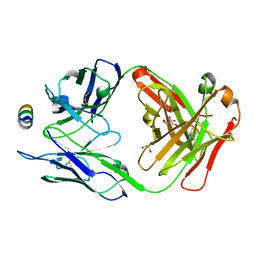 | | B6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M51
 
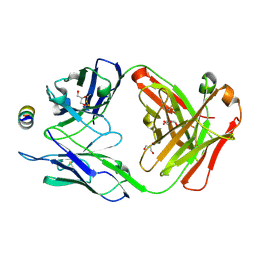 | | B6 Fab fragment bound to the OC43 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M55
 
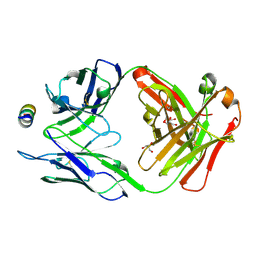 | | B6 Fab fragment bound to the MERS-CoV spike stem helix peptide | | Descriptor: | B6 antigen binding fragment (Fab) heavy chain, B6 antigen binding fragment (Fab) light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M5E
 
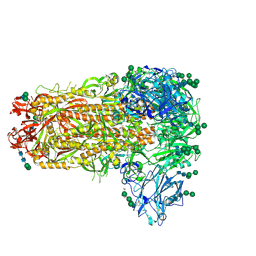 | |
7M52
 
 | | B6 Fab fragment bound to the HKU4 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5OVW
 
 | | Nanobody-bound BtuF, the vitamin B12 binding protein in Escherichia coli | | Descriptor: | GLYCEROL, Nanobody, Vitamin B12-binding protein | | Authors: | Mireku, S.A, Sauer, M.M, Glockshuber, R, Locher, K.P. | | Deposit date: | 2017-08-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD.
Sci Rep, 7, 2017
|
|
6GTZ
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound dimannoside Man(alpha1-3)Man in space group P21 | | Descriptor: | FimG protein, FimH protein, alpha-D-mannopyranose-(1-3)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTV
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound trimannose | | Descriptor: | DI(HYDROXYETHYL)ETHER, FimG protein, FimH protein, ... | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
4XOD
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 | | Descriptor: | FimG protein, FimH protein | | Authors: | Jakob, R.P, Sauer, M.M, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOC
 
 | | Crystal structure of the FimH lectin domain from E.coli F18 in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FimH protein, heptyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOE
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound heptyl alpha-D-mannopyrannoside | | Descriptor: | CACODYLATE ION, FimG protein, FimH protein, ... | | Authors: | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
7RNJ
 
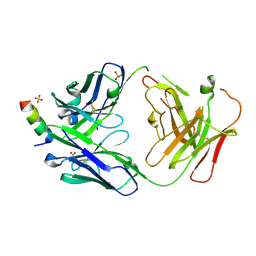 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
6GU0
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound dimannoside Man(alpha1-3)Man in space group P213 | | Descriptor: | FimG protein, FimH protein, SULFATE ION, ... | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTY
 
 | | Crystal structure of the FimH lectin domain from E.coli K12 in complex with the dimannoside Man(alpha1-6)Man | | Descriptor: | Type 1 fimbrin D-mannose specific adhesin, alpha-D-mannopyranose-(1-6)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTX
 
 | | Crystal structure of the FimH lectin domain from E.coli K12 in complex with the dimannoside Man(alpha1-2)Man | | Descriptor: | SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTW
 
 | | Crystal structure of the FimH lectin domain from E.coli F18 in complex with trimannose | | Descriptor: | CALCIUM ION, FimH protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
