6WQ3
 
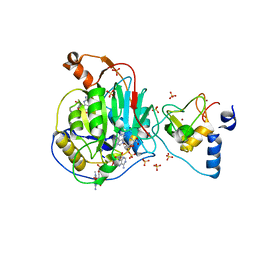 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-adenosyl-L-homocysteine. | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WRZ
 
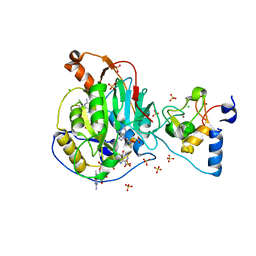 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 with 7-methyl-GpppA and S-adenosyl-L-homocysteine in the Active Site and Sulfates in the mRNA Binding Groove. | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XS4
 
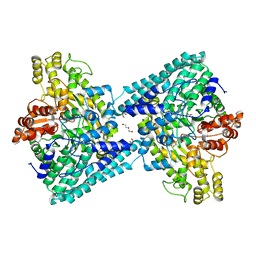 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
6XD8
 
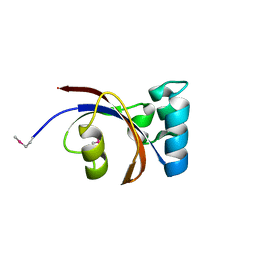 | | Crystal Structure of Peptidylprolyl Isomerase (PrsA) Fragment from Bacillus anthracis | | Descriptor: | Foldase protein PrsA 1 | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-10 | | Release date: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Peptidylprolyl Isomerase (PrsA) Fragment from Bacillus anthracis
To Be Published
|
|
8F4S
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8ER5
 
 | | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NlpC/P60 domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448.
To Be Published
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F8O
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids
To Be Published
|
|
8G22
 
 | | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae. | | Descriptor: | dTDP-4-dehydrorhamnose reductase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae.
To Be Published
|
|
8G1Y
 
 | | Crystal Structure of the Threonine Synthase from Streptococcus pneumoniae in complex with Pyridoxal 5-phosphate. | | Descriptor: | CHLORIDE ION, D-MALATE, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of the Threonine Synthase from Streptococcus pneumoniae in complex with Pyridoxal 5-phosphate.
To Be Published
|
|
8G28
 
 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | Descriptor: | ATPase, AAA family, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
7RKB
 
 | | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin | | Descriptor: | CHLORIDE ION, L-NEOPTERIN, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin.
To Be Published
|
|
7T1Q
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid | | Descriptor: | ACETATE ION, SUCCINIC ACID, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-02 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid.
To be Published
|
|
7SUA
 
 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
8D5H
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin
To Be Published
|
|
8D5I
 
 | | Crystal structure of dihydropteroate synthase H182G mutant (folP-SMZ_B27) from soil uncultured bacterium in complex with pteroic acid and pyrophosphate | | Descriptor: | PTEROIC ACID, PYROPHOSPHATE 2-, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Venkatesan, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of dihydropteroate synthase H182G mutant (folP-SMZ_B27) from soil uncultured bacterium in complex with pteroic acid
To Be Published
|
|
8D5G
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate | | Descriptor: | 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, CHLORIDE ION, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate
To Be Published
|
|
7RJJ
 
 | | Crystal Structure of the Peptidoglycan Binding Domain of the Outer Membrane Protein (OmpA) from Klebsiella pneumoniae with bound D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, OmpA family protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7RJ3
 
 | | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycogen accumulation regulator GarA | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis.
To Be Published
|
|
7RL8
 
 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
8SCD
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with reaction intermediate | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7MWA
 
 | | Crystal structure of 2-octaprenyl-6-methoxyphenol hydroxylase UbiH from Acinetobacter baumannii, apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-polyprenyl-6-methoxyphenol 4-hydroxylase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UbiH from Acinetobacter baumannii
To Be Published
|
|
