1UKU
 
 | | Crystal Structure of Pyrococcus horikoshii CutA1 Complexed with Cu2+ | | Descriptor: | COPPER (II) ION, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Yasutake, Y, Yao, M, Sakai, N, Tanaka, I, Tsumoto, K, Kumagai, I. | | Deposit date: | 2003-09-01 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
8H4M
 
 | | Crystal Structure of GTP-bound Irgb6_T95D mutant | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I
To Be Published
|
|
8H4O
 
 | | Crystal Structure of nucleotide-free Irgb6_T95D mutant | | Descriptor: | T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I.
Genes Cells, 29, 2024
|
|
8JRD
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
2F7L
 
 | | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase | | Descriptor: | 455aa long hypothetical phospho-sugar mutase, SULFATE ION | | Authors: | Kawamura, T, Sakai, N, Akutsu, J, Zhang, Z, Watanabe, N, Kawarabayashi, Y, Tanaka, I. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase
To be Published
|
|
4D4Z
 
 | | STRUCTURE OF HUMAN DEOXYHYPUSINE HYDROXYLASE in complex with glycerol | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYHYPUSINE HYDROXYLASE, FE (III) ION, ... | | Authors: | Han, Z, Sakai, N, Hilgenfeld, R. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Peroxo-Diiron(III) Intermediate of Deoxyhypusine Hydroxylase, an Oxygenase Involved in Hypusination.
Structure, 23, 2015
|
|
4D50
 
 | | Structure of human deoxyhypusine hydroxylase | | Descriptor: | DEOXYHYPUSINE HYDROXYLASE, FE (III) ION, GUANIDINE, ... | | Authors: | Han, Z, Sakai, N, Hilgenfeld, R. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Peroxo-Diiron(III) Intermediate of Deoxyhypusine Hydroxylase, an Oxygenase Involved in Hypusination.
Structure, 23, 2015
|
|
1IXL
 
 | | Crystal structure of uncharacterized protein PH1136 from Pyrococcus horikoshii | | Descriptor: | hypothetical protein PH1136 | | Authors: | Tajika, Y, Sakai, N, Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-06-27 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of conserved protein PH1136 from Pyrococcus horikoshii.
Proteins, 55, 2004
|
|
1IU8
 
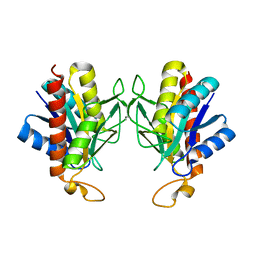 | | The X-ray Crystal Structure of Pyrrolidone-Carboxylate Peptidase from Hyperthermophilic Archaeon Pyrococcus horikoshii | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Sokabe, M, Kawamura, T, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-ray crystal structure of pyrrolidone-carboxylate peptidase from hyperthermophilic archaea Pyrococcus horikoshii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1IXK
 
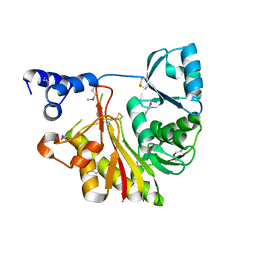 | | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | | Descriptor: | Methyltransferase | | Authors: | Ishikawa, I, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2002-06-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3A5P
 
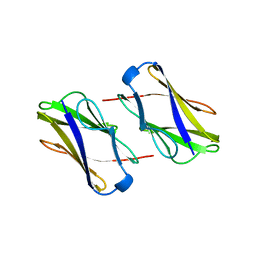 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
2ZDS
 
 | | Crystal Structure of SCO6571 from Streptomyces coelicolor A3(2) | | Descriptor: | Putative DNA-binding protein | | Authors: | Begum, P, Gao, Y.G, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SCO6571 from Streptomyces coelicolor A3(2).
Protein Pept.Lett., 15, 2008
|
|
7F0V
 
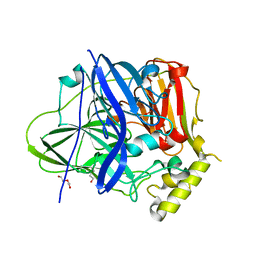 | | Structure of the M305I mutant of CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Kawano, T.K, Takata, S.T, Sakai, N.S, Imaizumi, R.I, Nakata, S.N, Takeshita, K.T, Yamashita, S.Y, Sakurai, T.S, Kataoka, K.K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structure of the M305I mutant of CueO
To Be Published
|
|
7F19
 
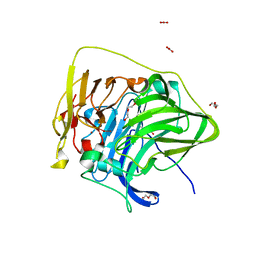 | | Structure of the G304E mutant of CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Takata, S.T, Kawano, T.K, Nakata, S.N, Takado, K.T, Imaizumi, R.I, Sakai, N.S, Takeshita, K.T, Yamashita, S.Y, Sakurai, T.S, Kataoka, K.K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure of the G304E mutant of CueO
To Be Published
|
|
7VES
 
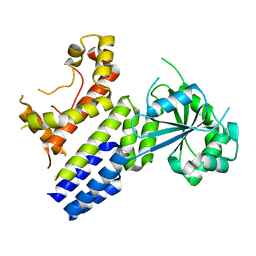 | | Crystal Structure of nucleotide-free Irgb6 | | Descriptor: | T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Sakai, N, Nitta, R. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of membrane recognition of Toxoplasma gondii vacuole by Irgb6.
Life Sci Alliance, 5, 2022
|
|
7VEX
 
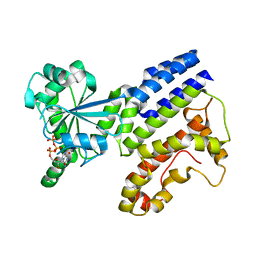 | | Crystal Structure of GTP-bound Irgb6 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Sakai, N, Nitta, R. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis of membrane recognition of Toxoplasma gondii vacuole by Irgb6.
Life Sci Alliance, 5, 2022
|
|
7VPC
 
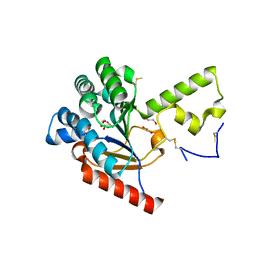 | | Neryl diphosphate synthase from Solanum lycopersicum | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Misawa, S, Takeshita, K, Sakai, N, Yamamoto, M, Kataoka, K, Nakayama, T, Takahashi, S, Yamashita, S. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based engineering of a short-chain cis-prenyltransferase to biosynthesize nonnatural all-cis-polyisoprenoids: molecular mechanisms for primer substrate recognition and ultimate product chain-length determination.
Febs J., 289, 2022
|
|
7VP0
 
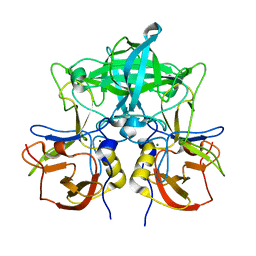 | | Crystal structure of P domain from norovirus GI.9 capsid protein. | | Descriptor: | MAGNESIUM ION, VP1 | | Authors: | Katsura, K, Sakai, N, Hasegawa, K, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lewis fucose is a key moiety for the recognition of histo-blood group antigens by GI.9 norovirus, as revealed by structural analysis.
Febs Open Bio, 12, 2022
|
|
1J2V
 
 | | Crystal Structure of CutA1 from Pyrococcus Horikoshii | | Descriptor: | 102AA long hypothetical periplasmic divalent cation tolerance protein CUTA | | Authors: | Tanaka, Y, Sakai, N, Yasutake, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
7X4N
 
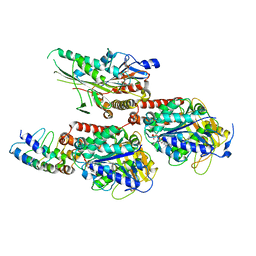 | | Crystal Structure of C. elegans kinesin-4 KLP-12 complexed with tubulin and DARPin | | Descriptor: | DARPin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Taguchi, S, Imasaki, T, Saijo-Hamano, Y, Sakai, N, Nitta, R. | | Deposit date: | 2022-03-03 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural model of microtubule dynamics inhibition by kinesin-4 from the crystal structure of KLP-12 -tubulin complex.
Elife, 11, 2022
|
|
1V30
 
 | | Crystal Structure Of Uncharacterized Protein PH0828 From Pyrococcus horikoshii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hypothetical UPF0131 protein PH0828 | | Authors: | Tajika, Y, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-10-21 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of hypothetical protein PH0828 from Pyrococcus horikoshii.
Proteins, 57, 2004
|
|
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
7COZ
 
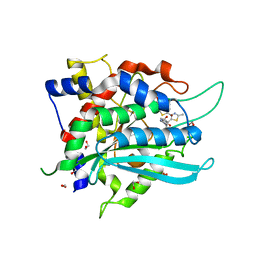 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-41 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(2-methyl-4-thiophen-2-yl-1,3-thiazol-5-yl)propanoyl]piperidine-4-carboxamide, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
1WLT
 
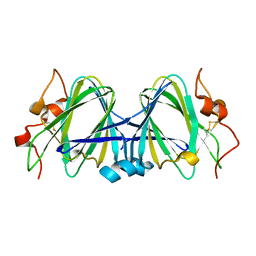 | | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue from Sulfolobus tokodaii | | Descriptor: | 176aa long hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Watanabe, N, Sakai, N, Zilian, Z, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue
to be published
|
|
1WU7
 
 | | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Tanaka, Y, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum
To be Published
|
|
