4UF6
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to an activating fragment of INO80G | | 分子名称: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, POLYUBIQUITIN-B, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | 著者 | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | 登録日 | 2014-12-23 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.69 Å) | | 主引用文献 | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UEL
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to the RPN13 DEUBAD domain | | 分子名称: | POLYUBIQUITIN-B, PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | 著者 | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | 登録日 | 2014-12-18 | | 公開日 | 2015-03-04 | | 最終更新日 | 2019-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UEM
 
 | | UCH-L5 in complex with the RPN13 DEUBAD domain | | 分子名称: | PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | 著者 | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | 登録日 | 2014-12-18 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UF5
 
 | | Crystal structure of UCH-L5 in complex with inhibitory fragment of INO80G | | 分子名称: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | 著者 | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | 登録日 | 2014-12-23 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
8FG6
 
 | |
2ASK
 
 | | Structure of human Artemin | | 分子名称: | SULFATE ION, artemin | | 著者 | Silvian, L, Jin, P, Carmillo, P, Boriack-Sjodin, P.A, Pelletier, C, Rushe, M, Gong, B.J, Sah, D, Pepinsky, B, Rossomando, A. | | 登録日 | 2005-08-23 | | 公開日 | 2006-06-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Artemin crystal structure reveals insights into heparan sulfate binding.
Biochemistry, 45, 2006
|
|
1P8T
 
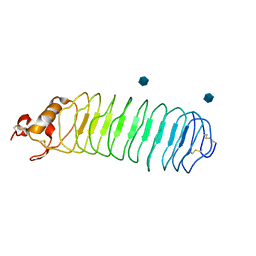 | | Crystal structure of Nogo-66 Receptor | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Reticulon 4 receptor | | 著者 | Barton, W.A, Liu, B.P, Tzvetkova, D, Jeffrey, P.D, Fournier, A.E, Sah, D, Cate, R, Strittmatter, S.M, Nikolov, D.B. | | 登録日 | 2003-05-07 | | 公開日 | 2003-05-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins
Embo J., 22, 2003
|
|
6WRX
 
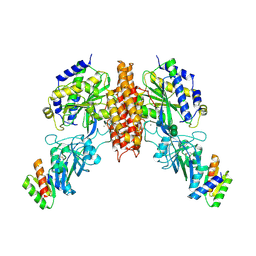 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRV
 
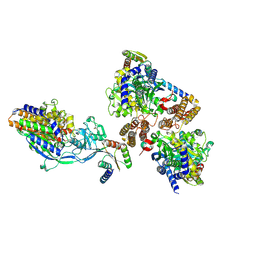 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | 著者 | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRW
 
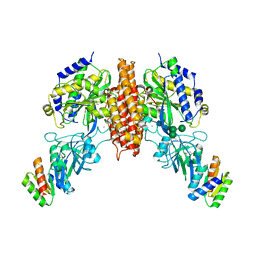 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | 著者 | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WMK
 
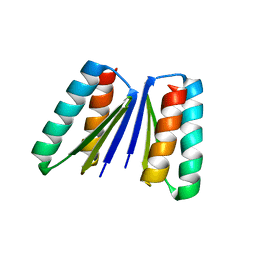 | | Crystal structure of beta sheet heterodimer LHD29 | | 分子名称: | Beta sheet heterodimer LHD29 - Chain A, Beta sheet heterodimer LHD29 - Chain B | | 著者 | Bera, A.K, Sahtoe, D.D, Kang, A, Sankaran, B, Baker, D. | | 登録日 | 2020-04-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
5KTF
 
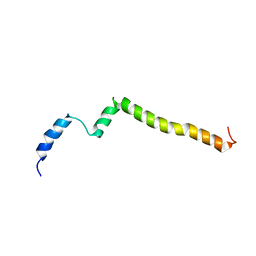 | | Structure of the C-terminal transmembrane domain of scavenger receptor BI (SR-BI) | | 分子名称: | Scavenger receptor class B member 1 | | 著者 | Chadwick, A.C, Peterson, F.C, Volkman, B.F, Sahoo, D. | | 登録日 | 2016-07-11 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif.
Structure, 25, 2017
|
|
3ZYK
 
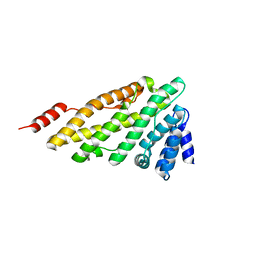 | | Structure of CALM (PICALM) ANTH domain | | 分子名称: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | 著者 | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | 登録日 | 2011-08-23 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYM
 
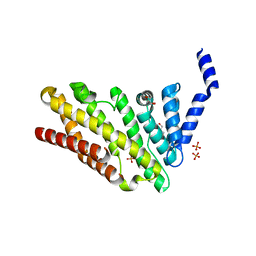 | | Structure of CALM (PICALM) in complex with VAMP8 | | 分子名称: | GLYCEROL, PHOSPHATE ION, PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN, ... | | 著者 | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | 登録日 | 2011-08-23 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYL
 
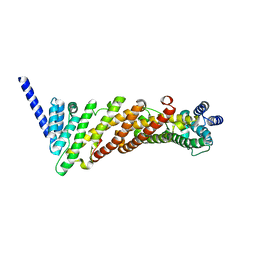 | | Structure of a truncated CALM (PICALM) ANTH domain | | 分子名称: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | 著者 | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | 登録日 | 2011-08-23 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
2NA2
 
 | |
7MWQ
 
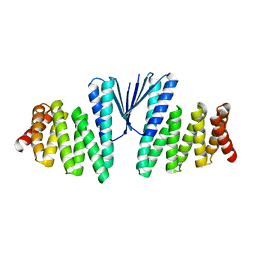 | | Structure of De Novo designed beta sheet heterodimer LHD29A53/B53 | | 分子名称: | LHD29A53, LHD29B53 | | 著者 | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | 登録日 | 2021-05-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7MWR
 
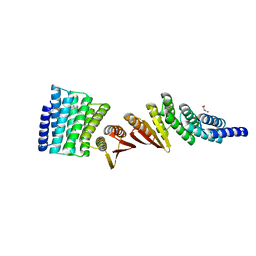 | | Structure of De Novo designed beta sheet heterodimer LHD101A53/B4 | | 分子名称: | LHD101A54, LHD101B4, MALONATE ION | | 著者 | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | 登録日 | 2021-05-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
1VRZ
 
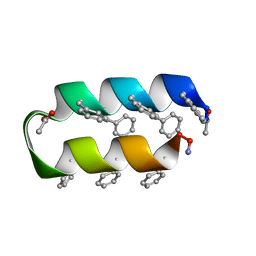 | | Helix turn helix motif | | 分子名称: | ACETATE ION, DE NOVO DESIGNED 21 RESIDUE PEPTIDE | | 著者 | Rudresh, Ramakumar, S, Ramagopal, U.A, Inai, Y, Sahal, D. | | 登録日 | 2005-10-14 | | 公開日 | 2005-11-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | De Novo Design and Characterization of a Helical Hairpin Eicosapeptide; Emergence of an Anion Receptor in the Linker Region.
Structure, 12, 2004
|
|
