5CBJ
 
 | |
3I54
 
 | | Crystal structure of MtbCRP in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, Crp/Fnr family | | Authors: | Reddy, M.C, Palaninathan, S.K, Bruning, J.B, Thurman, C, Smith, D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Mechanism of the Allosteric Transitions of Mycobacterium tuberculosis cAMP Receptor Protein.
J.Biol.Chem., 284, 2009
|
|
5ECV
 
 | |
5E9X
 
 | |
5DX7
 
 | |
5DRC
 
 | |
3I59
 
 | | Crystal structure of MtbCRP in complex with N6-cAMP | | Descriptor: | (2R)-N6-(1-Methyl-2-phenylethyl)adenosine-3',5'-cyclic monophosphate, (2S)-N6-(1-Methyl-2-phenylethyl)adenosine-3',5'-cyclic monophosphate, CHLORIDE ION, ... | | Authors: | Reddy, M.C, Palaninathan, S.K, Bruning, J.B, Thurman, C, Smith, D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insights into the Mechanism of the Allosteric Transitions of Mycobacterium tuberculosis cAMP Receptor Protein.
J.Biol.Chem., 284, 2009
|
|
5DRI
 
 | |
2H9I
 
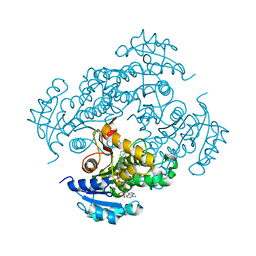 | | Mycobacterium tuberculosis InhA bound with ETH-NAD adduct | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], {(2R,3S,4R,5R)-5-[(4S)-3-(AMINOCARBONYL)-4-(2-ETHYLISONICOTINOYL)PYRIDIN-1(4H)-YL]-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL}METHYL [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of thioamide drug action against tuberculosis and leprosy.
J.Exp.Med., 204, 2007
|
|
5C9R
 
 | |
5CCZ
 
 | |
5C7V
 
 | |
5CAH
 
 | |
5CC6
 
 | |
5CC3
 
 | |
5CC7
 
 | |
7M7V
 
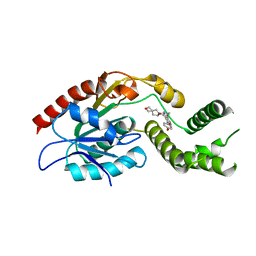 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with Compound 6 | | Descriptor: | 5-hydroxy-2-(4-hydroxyphenyl)-N-methyl-4-[(2-oxa-6-azaspiro[3.4]octan-6-yl)methyl]-1-benzofuran-3-carboxamide, Polyketide synthase Pks13 | | Authors: | Aggarwal, A, Sacchettini, J.C. | | Deposit date: | 2021-03-29 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Optimization of TAM16, a Benzofuran That Inhibits the Thioesterase Activity of Pks13; Evaluation toward a Preclinical Candidate for a Novel Antituberculosis Clinical Target.
J.Med.Chem., 65, 2022
|
|
1HKW
 
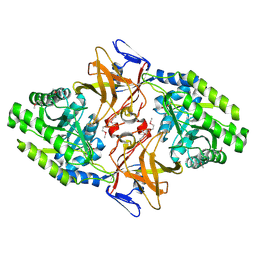 | | MYCOBACTERIUM DIAMINOPIMELATE DICARBOXYLASE (LysA) | | Descriptor: | DIAMINOPIMELATE DECARBOXYLASE, SULFATE ION | | Authors: | Gokulan, K, Rupp, B, Pavelka Jr, M.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-03-11 | | Release date: | 2003-03-18 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Diaminopimelate Decarboxylase, an Essential Enzyme in Bacterial Lysine Biosynthesis
J.Biol.Chem., 278, 2003
|
|
5CC5
 
 | |
1G5Z
 
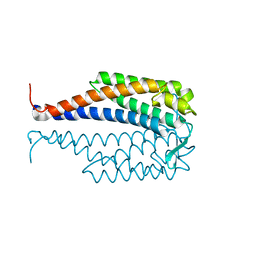 | | CRYSTAL STRUCTURE OF LYME DISEASE ANTIGEN OUTER SURFACE PROTEIN C (OSPC) FROM BORRELIA BURGDORFERI STRAIN N40 | | Descriptor: | OUTER SURFACE PROTEIN C | | Authors: | Eicken, C, Sharma, V, Klabunde, T, Owens, R.T, Pikas, D.S, Hook, M, Sacchettini, J.C. | | Deposit date: | 2000-11-02 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of Lyme disease antigen outer surface protein C from Borrelia burgdorferi.
J.Biol.Chem., 276, 2001
|
|
4GYQ
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase NDM-1, MAGNESIUM ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4HB7
 
 | | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50. | | Descriptor: | 1,2-ETHANEDIOL, Dihydropteroate synthase | | Authors: | Cuff, M.E, Holowicki, J, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50.
TO BE PUBLISHED
|
|
4H0D
 
 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae
To be Published
|
|
4HL2
 
 | | New Delhi Metallo-beta-Lactamase-1 1.05 A structure Complexed with Hydrolyzed Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 1.05 A structure Complexed with Hydrolyzed Ampicillin
To be Published
|
|
4HL1
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, CADMIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Ampicillin
To be Published
|
|
