5WW4
 
 | |
8OUO
 
 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 2024
|
|
8P2U
 
 | |
6HUE
 
 | | ParkinS65N | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | Deposit date: | 2018-10-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | Descriptor: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2023-05-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
5WNY
 
 | | DNA polymerase beta substrate complex with incoming 5-FdUTP | | Descriptor: | 2'-deoxy-5-fluorouridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
8JN6
 
 | | Cryo-EM structure of the small tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN3
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C (subparticle LLR-LRR) | | Descriptor: | Envelope protein, Human antibody DENV-115 heavy chain, Human antibody DENV-115 light chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN7
 
 | | Cryo-EM structure of the big tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN5
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 37 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein, Human antibody DENV-290 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN4
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 4 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein, Human antibody DENV-290 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN2
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 4 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein, Human antibody DENV-115 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN1
 
 | | Cryo-EM structure of dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein (Fragment), Human antibody DENV-115 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
5WW3
 
 | |
5WW9
 
 | |
8PJU
 
 | |
6FPX
 
 | | Structure of S. pombe Mmi1 in complex with 11-mer RNA | | Descriptor: | GLYCEROL, RNA (5'-R(P*UP*UP*UP*AP*AP*AP*CP*CP*UP*A)-3'), YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
8PJQ
 
 | |
8PJW
 
 | |
8PJZ
 
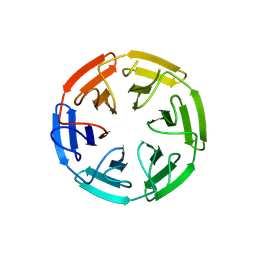 | |
8PJV
 
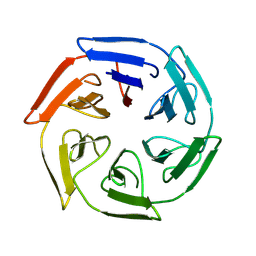 | |
8PJY
 
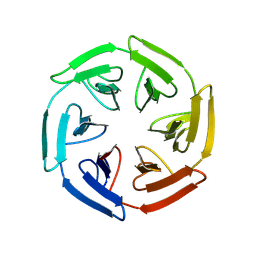 | |
8PJR
 
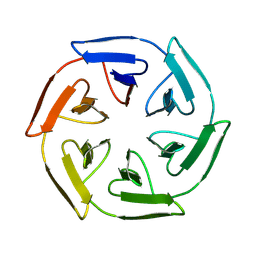 | |
8P7W
 
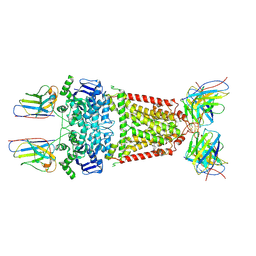 | | Structure of 5D3-Fab and nanobody(Nb8)-bound ABCG2 | | Descriptor: | 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ATP-binding cassette sub-family G member 2, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8P8J
 
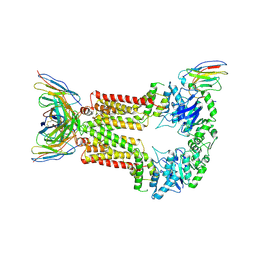 | | Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-06-01 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
