5MVW
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MW0
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
6TC7
 
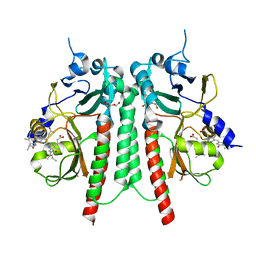 | | PAS-GAF bidomain of Glycine max phytochromeA | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
2GZX
 
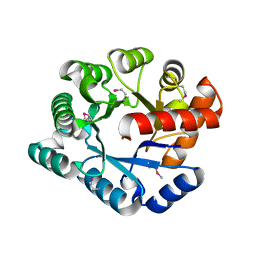 | | Crystal Structure of the TatD deoxyribonuclease MW0446 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR237. | | Descriptor: | NICKEL (II) ION, putative TatD related DNase | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Wang, D, Fang, Y, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the TatD deoxyribonuclease MW0446 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR237.
To be Published
|
|
5OMB
 
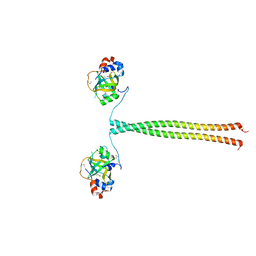 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
2GWJ
 
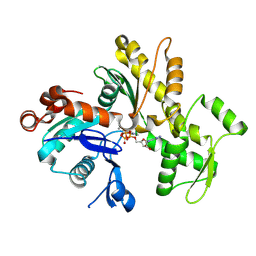 | |
5MVX
 
 | | Human DLL4 C2-EGF3 | | Descriptor: | Delta-like protein 4, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MW7
 
 | | Human Jagged2 C2-EGF3 | | Descriptor: | Protein jagged-2, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MWB
 
 | | Human Notch-2 EGF11-13 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 2, alpha-L-fucopyranose, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MWF
 
 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
6TNY
 
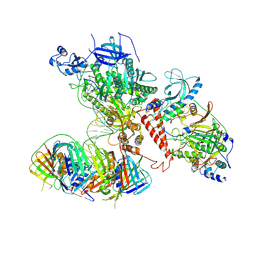 | | Processive human polymerase delta holoenzyme | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
4GWA
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6TL4
 
 | | Photosensory module (PAS-GAF-PHY) of Glycine max phyB | | Descriptor: | PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-12-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
2HF1
 
 | | Crystal structure of the putative Tetraacyldisaccharide-1-P 4-kinase from Chromobacterium violaceum. NESG target CvR39. | | Descriptor: | Tetraacyldisaccharide-1-P 4-kinase, ZINC ION | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Chen, C.X, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative Tetraacyldisaccharide-1-P 4-kinase from Chromobacterium
violaceum.
To be Published
|
|
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | Descriptor: | DNA damage checkpoint protein LCD1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
2HLR
 
 | |
3BC2
 
 | |
2FEJ
 
 | | Solution structure of human p53 DNA binding domain. | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Perez-Canadillas, J.M, Tidow, H, Freund, S.M, Rutherford, T.J, Ang, H.C, Fersht, A.R. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of p53 core domain: Structural basis for its instability
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GWK
 
 | |
4HAP
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2I18
 
 | |
2FAH
 
 | | The structure of mitochondrial PEPCK, Complex with Mn and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MALONIC ACID, MANGANESE (II) ION, ... | | Authors: | Holyoak, T, Sullivan, S.M, Nowak, T. | | Deposit date: | 2005-12-07 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights into the Mechanism of PEPCK Catalysis
Biochemistry, 45, 2006
|
|
2FAF
 
 | | The structure of chicken mitochondrial PEPCK. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MANGANESE (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Holyoak, T, Sullivan, S.M, Nowak, T. | | Deposit date: | 2005-12-07 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Mechanism of PEPCK Catalysis
Biochemistry, 45, 2006
|
|
2FM2
 
 | | HCV NS3-4A protease domain complexed with a ketoamide inhibitor, SCH446211 | | Descriptor: | BETA-MERCAPTOETHANOL, NS3 protease/helicase, NS4a protein, ... | | Authors: | Yi, M, Tong, X, Skelton, A, Chase, R, Chen, T, Prongay, A, Bogen, S.L, Saksena, A.K, Njoroge, F.G, Veselenak, R.L, Pyles, R.B, Bourne, N, Malcolm, B.A, Lemon, S.M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations
J.Biol.Chem., 281, 2006
|
|
2F20
 
 | | X-ray Crystal Structure of Protein BT_1218 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR8. | | Descriptor: | conserved hypothetical protein, with conserved domain | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Forouhar, F, Xiao, R, Ma, L.-C, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Q8A8E9_BACTIN hypothetical protein from Bacteroides thetaiotaomicron.
To be Published
|
|
