5GG5
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hydrolase, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GG8
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP, 8-oxo-dGMP and pyrophosphate (II) | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GG9
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-GTP, 8-oxo-GMP and pyrophosphate | | Descriptor: | 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, Hydrolase, NUDIX family protein, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGA
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-GDP, 8-oxo-GMP and pyrophosphate | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3O3F
 
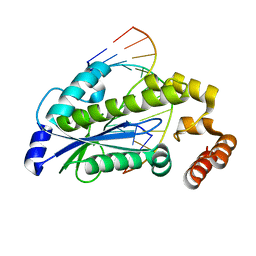 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and magnesium ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
5GGD
 
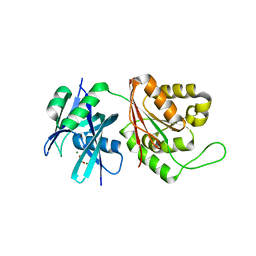 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with phosphate and magnesium ions (excess magnesium, II) | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3O8Z
 
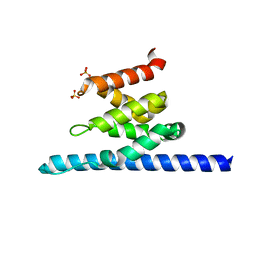 | | Crystal structure of Spn1 (Iws1) core domain | | Descriptor: | SULFATE ION, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
3O3H
 
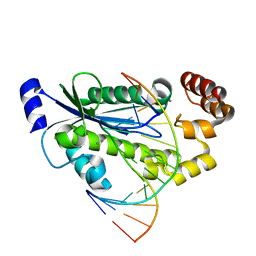 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and manganese ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
3NWV
 
 | | Human cytochrome c G41S | | Descriptor: | Cytochrome c, HEME C | | Authors: | Fagerlund, R.D, Wilbanks, S.M. | | Deposit date: | 2010-07-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Proapoptotic G41S Mutation to Human Cytochrome c Alters the Heme Electronic Structure and Increases the Electron Self-Exchange Rate.
J.Am.Chem.Soc., 133, 2011
|
|
5KEQ
 
 | | High resolution cryo-EM maps of Human papillomavirus 16 reveal L2 location and heparin-induced conformational changes | | Descriptor: | Major capsid protein L1 | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryoelectron Microscopy Maps of Human Papillomavirus 16 Reveal L2 Densities and Heparin Binding Site.
Structure, 25, 2017
|
|
2X1K
 
 | |
8FTU
 
 | | Crystal structure of the SNARE Use1 bound to Dsl1 complex subunits Sec39 and Dsl1, Revised Use1 structure | | Descriptor: | Protein transport protein DSL1, Protein transport protein SEC39, Vesicle transport protein USE1 | | Authors: | Travis, S.M, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5.73 Å) | | Cite: | Structure of a membrane tethering complex incorporating multiple SNAREs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
1HFO
 
 | |
1IJD
 
 | | Crystallographic Structure of the LH3 Complex from Rhodopseudomonas acidophila strain 7050 | | Descriptor: | BACTERIOCHLOROPHYLL A, LIGHT-HARVESTING PROTEIN B-800/820, ALPHA CHAIN, ... | | Authors: | McLuskey, K, Prince, S.M, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2001-04-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystallographic structure of the B800-820 LH3 light-harvesting complex from the purple bacteria Rhodopseudomonas acidophila strain 7050.
Biochemistry, 40, 2001
|
|
8SLO
 
 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
2Y4R
 
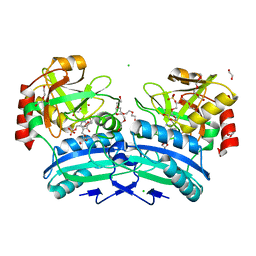 | | CRYSTAL STRUCTURE OF 4-AMINO-4-DEOXYCHORISMATE LYASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-4-DEOXYCHORISMATE LYASE, CHLORIDE ION, ... | | Authors: | O'Rourke, P.E.F, Eadsforth, T.C, Fyfe, P.K, Shepard, S.M, Agacan, M, Hunter, W.N. | | Deposit date: | 2011-01-10 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pseudomonas Aeruginosa 4-Amino-4-Deoxychorismate Lyase: Spatial Conservation of an Active Site Tyrosine and Classification of Two Types of Enzyme.
Plos One, 6, 2011
|
|
1WYZ
 
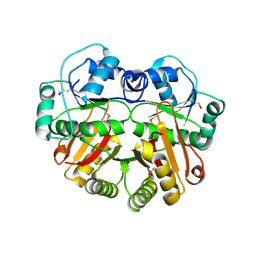 | | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28 | | Descriptor: | putative S-adenosylmethionine-dependent methyltransferase | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-21 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28
To be Published
|
|
1XDV
 
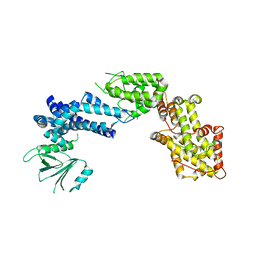 | | Experimentally Phased Structure of Human the Son of Sevenless protein at 4.1 Ang. | | Descriptor: | Son of sevenless protein homolog 1 | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1GNP
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID 3'-O-(N-METHYLANTHRANILOYL-2'-DEOXYGUANYLATE ESTER | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
5KUX
 
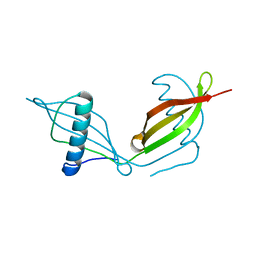 | |
5KUY
 
 | |
1X9Q
 
 | | 4m5.3 anti-fluorescein single chain antibody fragment (scFv) | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 4m5.3 anti-fluorescein single chain antibody fragment, ACETATE ION | | Authors: | Midelfort, K.S, Hernandez, H.H, Lippow, S.M, Tidor, B, Drennan, C.L, Wittrup, K.D. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substantial energetic improvement with minimal structural perturbation in a high affinity mutant antibody
J.Mol.Biol., 343, 2004
|
|
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
2X1J
 
 | |
