3MGK
 
 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3PBK
 
 | |
8SZJ
 
 | | Human glutaminase C (Y466W) with L-Gln and Pi, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-29 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
3MMZ
 
 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | Authors: | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
8T0Z
 
 | | Human liver-type glutaminase (K253A) with L-Gln, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
3PDM
 
 | | Hibiscus Latent Singapore virus | | Descriptor: | Coat protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Tewary, S.K, Wong, S.M, Swaminathan, K. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | FIBER DIFFRACTION (3.5 Å) | | Cite: | Structure of Hibiscus latent Singapore virus by fiber diffraction: A non-conserved His122 contributes to coat protein stability
J.Mol.Biol., 2010
|
|
3PDW
 
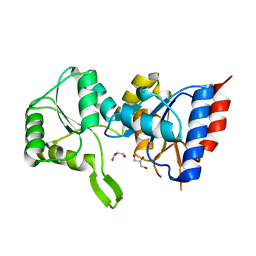 | | Crystal structure of putative p-nitrophenyl phosphatase from Bacillus subtilis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized hydrolase yutF | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Crystal structure of putative p-nitrophenyl phosphatase from Bacillus subtilis
To be Published
|
|
3MJS
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | (2S)-2-hydroxybutanedioic acid, AmphB, D-MALATE, ... | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
8SZL
 
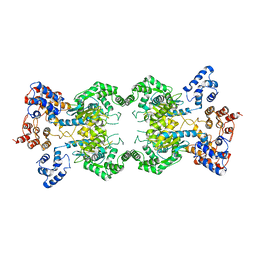 | | Human liver-type glutaminase (Apo form) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
3PBM
 
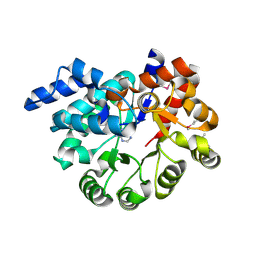 | |
3MF4
 
 | | Crystal structure of putative two-component system response regulator/ggdef domain protein | | Descriptor: | MAGNESIUM ION, Two-component system response regulator/GGDEF domain protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative two-component system response regulator/ggdef domain protein
To be Published
|
|
1BY8
 
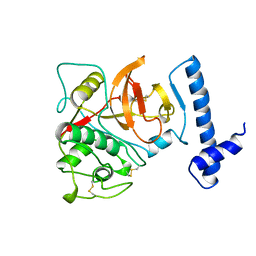 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
3MJV
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
3M0G
 
 | | CRYSTAL STRUCTURE OF putative farnesyl diphosphate synthase from Rhodobacter capsulatus | | Descriptor: | Farnesyl diphosphate synthase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative farnesyl diphosphate synthase from Rhodobacter capsulatus
To be Published
|
|
3M6I
 
 | | L-arabinitol 4-dehydrogenase | | Descriptor: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
3MWC
 
 | | Crystal structure of probable o-succinylbenzoic acid synthetase from kosmotoga olearia | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-05 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of O-Succinylbenzoic Acid Synthetase from Kosmotoga Olearia
To be Published
|
|
1BO4
 
 | | CRYSTAL STRUCTURE OF A GCN5-RELATED N-ACETYLTRANSFERASE: SERRATIA MARESCENS AMINOGLYCOSIDE 3-N-ACETYLTRANSFERASE | | Descriptor: | COENZYME A, PROTEIN (SERRATIA MARCESCENS AMINOGLYCOSIDE-3-N-ACETYLTRANSFERASE), SPERMIDINE | | Authors: | Wolf, E, Vassilev, A, Makino, Y, Sali, A, Nakatani, Y, Burley, S.K. | | Deposit date: | 1998-08-08 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BVA
 
 | | MANGANESE BINDING MUTANT IN CYTOCHROME C PEROXIDASE | | Descriptor: | MANGANESE (II) ION, PROTEIN (CYTOCHROME C PEROXIDASE), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Rational design of a functional metalloenzyme: introduction of a site for manganese binding and oxidation into a heme peroxidase.
Biochemistry, 37, 1998
|
|
3N28
 
 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | Descriptor: | Phosphoserine phosphatase, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
3MZN
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 | | Descriptor: | ACETATE ION, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
8SF7
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain MEC17 | | Descriptor: | CFAM166A, CFAM166B, CFAM166C, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effect of alpha-tubulin acetylation on the doublet microtubule structure.
Elife, 12, 2024
|
|
3QQX
 
 | | Reduced Native Intermediate of the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CueO E506D Mutant: Crystal Structure of Reduced Native Intermediate, Kinetics, and Impairment of Product Release
To be Published
|
|
3QRC
 
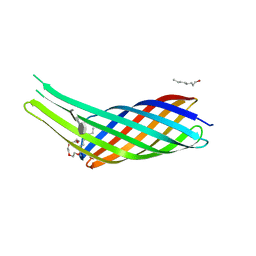 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis, in complex with the heparin analogue sucrose octasulfate | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
1BXD
 
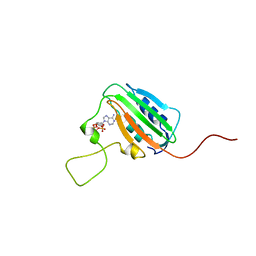 | | NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (OSMOLARITY SENSOR PROTEIN (ENVZ)) | | Authors: | Tanaka, T, Saha, S.K, Tomomori, C, Ishima, R, Liu, D, Tong, K.I, Park, H, Dutta, R, Qin, L, Swindells, M.B, Yamazaki, T, Ono, A.M, Kainosho, M, Inouye, M, Ikura, M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ.
Nature, 396, 1998
|
|
3N05
 
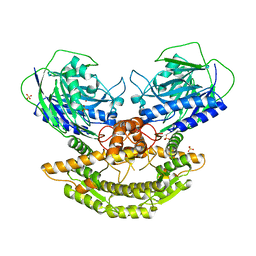 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM STREPTOMYCES AVERMITILIS | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Nh3-Dependent Nad+ Synthetase from Streptomyces Avermitilis
To be Published
|
|
