5QD1
 
 | | Crystal structure of BACE complex with BMC011 | | 分子名称: | (10S,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-7-oxa-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6T5M
 
 | |
7QV7
 
 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | 分子名称: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | 著者 | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | 登録日 | 2022-01-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
5QBX
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | (2S)-1-[4-(2-methoxyphenyl)piperidin-1-yl]-3-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propan-2-ol, Cathepsin S | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCE
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | Cathepsin S, N-benzyl-1-{2-chloro-5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCT
 
 | | Crystal structure of BACE complex with BMC001 | | 分子名称: | (2R,4S)-N-butyl-4-[(4S,6R)-16-ethoxy-12-ethyl-6-methyl-2,13-dioxo-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-trien-4-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1, PHOSPHATE ION | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD4
 
 | | Crystal structure of BACE complex with BMC023 | | 分子名称: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.112 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
7U36
 
 | | Crystal structure of human GSK3B in complex with ARN1484 | | 分子名称: | (3S)-1-[(2-fluorophenoxy)acetyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | 著者 | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2022-02-25 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U2Z
 
 | | Crystal structure of human GSK3B in complex with G12 | | 分子名称: | (3R)-1-[3-(2-fluorophenyl)propanoyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | 著者 | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2022-02-25 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U31
 
 | | Crystal structure of human GSK3B in complex with G5 | | 分子名称: | 5-(4-fluorophenyl)-4-[1-(methanesulfonyl)azetidin-3-yl]pyrimidin-2-amine, CHLORIDE ION, Glycogen synthase kinase-3 beta | | 著者 | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2022-02-25 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U33
 
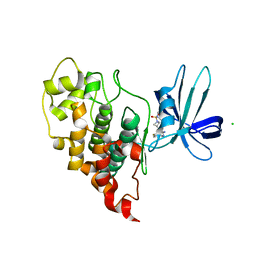 | | Crystal structure of human GSK3B in complex with ARN9133 | | 分子名称: | 3-[2-amino-5-(4-fluorophenyl)pyrimidin-4-yl]-N,N-dimethylazetidine-1-sulfonamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | 著者 | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2022-02-25 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
6T4V
 
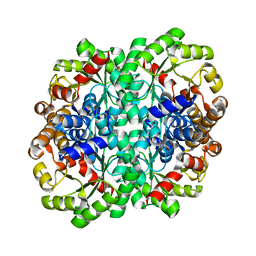 | |
7U58
 
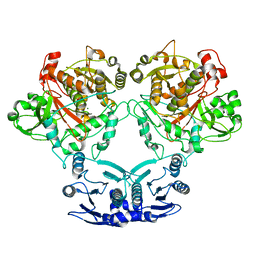 | |
6FMP
 
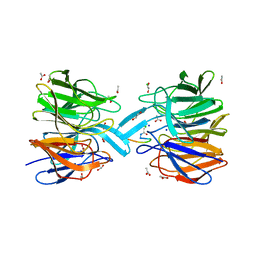 | | Keap1 - peptide complex | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, ACY-ASP-GLU-GLU-THR-GLY-GLU-PHE, ... | | 著者 | Talapatra, S.K, Kozielski, F, Wells, G, Georgakopoulos, N.D. | | 登録日 | 2018-02-01 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Modified Peptide Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction Incorporating Unnatural Amino Acids.
Chembiochem, 19, 2018
|
|
6MP1
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with the mutant TRP1-K8 peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TRP1-K8 peptide, Beta-2-microglobulin,H-2 class I histocompatibility antigen, ... | | 著者 | Clancy-Thompson, E, Devlin, C.A, Birnbaum, M.E, Dougan, S.K. | | 登録日 | 2018-10-05 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.211 Å) | | 主引用文献 | Altered Binding of Tumor Antigenic Peptides to MHC Class I Affects CD8+T Cell-Effector Responses.
Cancer Immunol Res, 6, 2018
|
|
3B89
 
 | |
3BGE
 
 | | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae | | 分子名称: | Predicted ATPase, SULFATE ION | | 著者 | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Meyer, A.J, Toro, R, Freeman, J, Adams, J, Koss, J, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-11-26 | | 公開日 | 2008-01-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae.
To be Published
|
|
2IMH
 
 | | Crystal structure of protein SPO2555 from Silicibacter pomeroyi, Pfam DUF1028 | | 分子名称: | Hypothetical protein UNP Q5LQD5_SILPO | | 著者 | Bonanno, J.B, Dickey, M, Bain, K.T, Slocombe, A, Ozyurt, S, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-10-04 | | 公開日 | 2006-10-17 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal structure of hypothetical protein from
Silicibacter pomeroyi
To be Published
|
|
2IMR
 
 | |
3B40
 
 | | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa | | 分子名称: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Bonanno, J.B, Patskovsky, Y, Dickey, M, Bain, K.T, Mendoza, M, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-10-23 | | 公開日 | 2007-11-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa.
To be Published
|
|
5QC7
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S, DIMETHYL SULFOXIDE, ... | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
6TNM
 
 | | E. coli aerobic trifunctional enzyme subunit-alpha | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Fatty acid oxidation complex subunit alpha, GLYCEROL, ... | | 著者 | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | 登録日 | 2019-12-09 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Insights into the stability and substrate specificity of the E. coli aerobic beta-oxidation trifunctional enzyme complex.
J.Struct.Biol., 210, 2020
|
|
5QCZ
 
 | | Crystal structure of BACE complex with BMC015 | | 分子名称: | (4S)-4-{(S)-hydroxy[(3R,6R)-6-(methoxymethyl)morpholin-3-yl]methyl}-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
2K3V
 
 | | Solution Structure of a Tetrahaem Cytochrome from Shewanella Frigidimarina | | 分子名称: | HEME C, Tetraheme cytochrome c-type | | 著者 | Paixao, V.B, Turner, D.L, Salgueiro, C.A, Brennan, L, Reid, G.A, Chapman, S.K. | | 登録日 | 2008-05-19 | | 公開日 | 2009-03-31 | | 最終更新日 | 2019-10-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a tetraheme cytochrome from Shewanella frigidimarina reveals a novel family structural motif
Biochemistry, 47, 2008
|
|
6G6Z
 
 | | Eg5-inhibitor complex | | 分子名称: | (~{N}~{Z})-~{N}-[(5~{S})-4-ethanoyl-5-methyl-5-phenyl-1,3,4-thiadiazolidin-2-ylidene]ethanamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | 著者 | Talapatra, S.K, Kozielski, F, Tham, C.L. | | 登録日 | 2018-04-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the Eg5 - K858 complex and implications for structure-based design of thiadiazole-containing inhibitors.
Eur J Med Chem, 156, 2018
|
|
